4. Exploratory Data Analysis#
Now we get to start actual data science!
4.1. This week: Exploratory Data Analysis#
How to summarize data
Interpretting summaries
Visualizing data
interpretting summaries
4.1.1. Summarizing and Visualizing Data are very important#
People cannot interpret high dimensional or large samples quickly
Important in EDA to help you make decisions about the rest of your analysis
Important in how you report your results
Summaries are similar calculations to performance metrics we will see later
visualizations are often essential in debugging models
THEREFORE
You have a lot of chances to earn summarize and visualize
we will be picky when we assess if you earned them or not
import pandas as pd
Today we will work with a new dataset about Carbon emissions
carbon_data_url = 'https://github.com/rfordatascience/tidytuesday/raw/master/data/2024/2024-05-21/emissions.csv'
carbon_df = pd.read_csv(carbon_data_url)
---------------------------------------------------------------------------
HTTPError Traceback (most recent call last)
Cell In[3], line 1
----> 1 carbon_df = pd.read_csv(carbon_data_url)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/io/parsers/readers.py:912, in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, date_format, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options, dtype_backend)
899 kwds_defaults = _refine_defaults_read(
900 dialect,
901 delimiter,
(...)
908 dtype_backend=dtype_backend,
909 )
910 kwds.update(kwds_defaults)
--> 912 return _read(filepath_or_buffer, kwds)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/io/parsers/readers.py:577, in _read(filepath_or_buffer, kwds)
574 _validate_names(kwds.get("names", None))
576 # Create the parser.
--> 577 parser = TextFileReader(filepath_or_buffer, **kwds)
579 if chunksize or iterator:
580 return parser
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/io/parsers/readers.py:1407, in TextFileReader.__init__(self, f, engine, **kwds)
1404 self.options["has_index_names"] = kwds["has_index_names"]
1406 self.handles: IOHandles | None = None
-> 1407 self._engine = self._make_engine(f, self.engine)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/io/parsers/readers.py:1661, in TextFileReader._make_engine(self, f, engine)
1659 if "b" not in mode:
1660 mode += "b"
-> 1661 self.handles = get_handle(
1662 f,
1663 mode,
1664 encoding=self.options.get("encoding", None),
1665 compression=self.options.get("compression", None),
1666 memory_map=self.options.get("memory_map", False),
1667 is_text=is_text,
1668 errors=self.options.get("encoding_errors", "strict"),
1669 storage_options=self.options.get("storage_options", None),
1670 )
1671 assert self.handles is not None
1672 f = self.handles.handle
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/io/common.py:716, in get_handle(path_or_buf, mode, encoding, compression, memory_map, is_text, errors, storage_options)
713 codecs.lookup_error(errors)
715 # open URLs
--> 716 ioargs = _get_filepath_or_buffer(
717 path_or_buf,
718 encoding=encoding,
719 compression=compression,
720 mode=mode,
721 storage_options=storage_options,
722 )
724 handle = ioargs.filepath_or_buffer
725 handles: list[BaseBuffer]
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/io/common.py:368, in _get_filepath_or_buffer(filepath_or_buffer, encoding, compression, mode, storage_options)
366 # assuming storage_options is to be interpreted as headers
367 req_info = urllib.request.Request(filepath_or_buffer, headers=storage_options)
--> 368 with urlopen(req_info) as req:
369 content_encoding = req.headers.get("Content-Encoding", None)
370 if content_encoding == "gzip":
371 # Override compression based on Content-Encoding header
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/site-packages/pandas/io/common.py:270, in urlopen(*args, **kwargs)
264 """
265 Lazy-import wrapper for stdlib urlopen, as that imports a big chunk of
266 the stdlib.
267 """
268 import urllib.request
--> 270 return urllib.request.urlopen(*args, **kwargs)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/urllib/request.py:222, in urlopen(url, data, timeout, cafile, capath, cadefault, context)
220 else:
221 opener = _opener
--> 222 return opener.open(url, data, timeout)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/urllib/request.py:531, in OpenerDirector.open(self, fullurl, data, timeout)
529 for processor in self.process_response.get(protocol, []):
530 meth = getattr(processor, meth_name)
--> 531 response = meth(req, response)
533 return response
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/urllib/request.py:640, in HTTPErrorProcessor.http_response(self, request, response)
637 # According to RFC 2616, "2xx" code indicates that the client's
638 # request was successfully received, understood, and accepted.
639 if not (200 <= code < 300):
--> 640 response = self.parent.error(
641 'http', request, response, code, msg, hdrs)
643 return response
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/urllib/request.py:569, in OpenerDirector.error(self, proto, *args)
567 if http_err:
568 args = (dict, 'default', 'http_error_default') + orig_args
--> 569 return self._call_chain(*args)
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/urllib/request.py:502, in OpenerDirector._call_chain(self, chain, kind, meth_name, *args)
500 for handler in handlers:
501 func = getattr(handler, meth_name)
--> 502 result = func(*args)
503 if result is not None:
504 return result
File /opt/hostedtoolcache/Python/3.8.18/x64/lib/python3.8/urllib/request.py:649, in HTTPDefaultErrorHandler.http_error_default(self, req, fp, code, msg, hdrs)
648 def http_error_default(self, req, fp, code, msg, hdrs):
--> 649 raise HTTPError(req.full_url, code, msg, hdrs, fp)
HTTPError: HTTP Error 404: Not Found
carbon_df.head()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[4], line 1
----> 1 carbon_df.head()
NameError: name 'carbon_df' is not defined
In this case, head shows that these rows are very similar because it is sorted. Tail would probably be similar, so we will try sample to get a random subset.
carbon_df.sample(5)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[5], line 1
----> 1 carbon_df.sample(5)
NameError: name 'carbon_df' is not defined
4.2. Describing a Dataset#
So far, we’ve loaded data in a few different ways and then we’ve examined DataFrames as a data structure, looking at what different attributes they have and what some of the methods are, and how to get data into them.
The describe method provides us with a set of summary statistics that broadly
carbon_df.describe()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[6], line 1
----> 1 carbon_df.describe()
NameError: name 'carbon_df' is not defined
And these all give us a sense of the values and the distribution or spread fo the data in each column.
4.2.1. Individual statistics#
We can also extract each of the statistics that the describe method calculates individually, by name.
carbon_df.max()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[7], line 1
----> 1 carbon_df.max()
NameError: name 'carbon_df' is not defined
4.2.2. Understanding Quantiles#
The 50% has another more common name: the median. It means 50% of the data are lower (and higher) than this value.
The quantiles
are tricky, we cannot just .25%() to get the 25% percentile, we have to use the
quantile method and pass it a value between 0 and 1.
carbon_df['production_value'].quantile(.25)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[8], line 1
----> 1 carbon_df['production_value'].quantile(.25)
NameError: name 'carbon_df' is not defined
carbon_df['production_value'].quantile(.8)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[9], line 1
----> 1 carbon_df['production_value'].quantile(.8)
NameError: name 'carbon_df' is not defined
4.3. Individual variable#
We can use the descriptive statistics on individual columns as well
4.4. What is the average total emissions in this dataset?#
carbon_df['total_emissions_MtCO2e'].mean()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[10], line 1
----> 1 carbon_df['total_emissions_MtCO2e'].mean()
NameError: name 'carbon_df' is not defined
4.5. Working with categorical data#
There are different columns in the describe than the the whole dataset:
carbon_df.columns
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[11], line 1
----> 1 carbon_df.columns
NameError: name 'carbon_df' is not defined
So far, the stats above are only for numerical features.
We can get the prevalence of each one with value_counts, this can also allow us to answer certain questions. Let’s try a few.
4.5.1. What is the most common parent type?#
carbon_df['parent_type'].value_counts()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[12], line 1
----> 1 carbon_df['parent_type'].value_counts()
NameError: name 'carbon_df' is not defined
Show code cell source
from myst_nb import glue
most_common_ptype = carbon_df['parent_type'].value_counts().idxmax()
glue('max_parent',most_common_ptype)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[13], line 2
1 from myst_nb import glue
----> 2 most_common_ptype = carbon_df['parent_type'].value_counts().idxmax()
3 glue('max_parent',most_common_ptype)
NameError: name 'carbon_df' is not defined
This shows us that the most common one is
We also notice that the data is relatively balanced enough that we can make good comparisons from the overall output.
For the specific question, we could instead use the mode method (the most frequent value).
carbon_df['parent_type'].mode()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[14], line 1
----> 1 carbon_df['parent_type'].mode()
NameError: name 'carbon_df' is not defined
4.5.2. Is it reasonable to compare and use all of the the commodity types in the data#
To do this, we again look at the value counts:
carbon_df['commodity'].value_counts()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[15], line 1
----> 1 carbon_df['commodity'].value_counts()
NameError: name 'carbon_df' is not defined
Here, we note that the top few are a lot more similar and the smallest one is a lot lower. Here we might drop only the bottom couple or keep them call becaus for a lot of things 200 measurements is enough.
Another choice is that we could combine the different types of coals together. We can do this by adding a column.
carbon_df['commodity_simple'] = carbon_df['commodity'].apply(lambda s: s if not('Coal' in s) else 'Coal')
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[16], line 1
----> 1 carbon_df['commodity_simple'] = carbon_df['commodity'].apply(lambda s: s if not('Coal' in s) else 'Coal')
NameError: name 'carbon_df' is not defined
The way this works is:
carbon_df['commodity']picks out the column of interestthe
.applymethod applies a function (that we pass to it) to all of the elements of a series (or to the rows or columns of a DataFrame)lambda s: s if not('Coal' in s) else 'Coal'is a lambda function that returns the same value if Col is not in it or onlyCoalif it isassigns values to a column
'commodity_simple'carbon_df['commodity_simple'] =. It also creates that column since it did not exist
4.5.3. understanding lambda functions#
To better understand the lambda function that we used above, let’s assign it to a variable.
coal_strip = lambda s: s if not('Coal' in s) else 'Coal'
First we will inspect it a little:
type(coal_strip)
function
We can see it is a function here
Now we can make calls to it, like any other functions
coal_strip('Metallurgical Coal')
'Coal'
this works and now we test the other case:
coal_strip('naything else')
'naything else'
4.5.4. How a column is added#
Now we can see a sample of the DataFrame again to see how the apply method works.
carbon_df.sample(5)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[21], line 1
----> 1 carbon_df.sample(5)
NameError: name 'carbon_df' is not defined
We can further examine this column:
carbon_df['commodity_simple'].value_counts()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[22], line 1
----> 1 carbon_df['commodity_simple'].value_counts()
NameError: name 'carbon_df' is not defined
Now the numbers are more similar, we might want to drop the cement, we can do that using a mask that we saw last week.
4.6. Split-Apply-Combine#
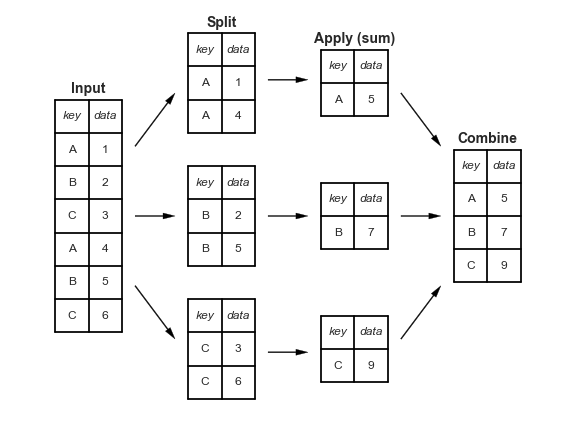
see it in action on pandas tutor
4.6.1. Which commodity type has the highest average emissions?#
To do this we group by the categorical value, the new simplified commodity column we created, then we pick out thee emissions column, and take the average.
carbon_df.groupby('commodity_simple')['total_emissions_MtCO2e'].mean()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[23], line 1
----> 1 carbon_df.groupby('commodity_simple')['total_emissions_MtCO2e'].mean()
NameError: name 'carbon_df' is not defined
4.6.2. How apply works#
We can use apply with any function, that we write in advance, that comes from a library, or is built in. Above, we used a lambda we defined on the fly, but here we can make it first.
def first_chars(s):
return s[:5]
carbon_df['commodity'].apply(first_chars)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[24], line 4
1 def first_chars(s):
2 return s[:5]
----> 4 carbon_df['commodity'].apply(first_chars)
NameError: name 'carbon_df' is not defined
4.6.3. Grouping Multiple times#
we can pass a list to group by two values
carbon_df.groupby(['commodity_simple','parent_type'])['total_emissions_MtCO2e'].mean()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[25], line 1
----> 1 carbon_df.groupby(['commodity_simple','parent_type'])['total_emissions_MtCO2e'].mean()
NameError: name 'carbon_df' is not defined
This is one Series, but is has a multi-index. We can check the type
em_by_comm_parent = carbon_df.groupby(['commodity_simple','parent_type'])['total_emissions_MtCO2e'].mean()
type(em_by_comm_parent)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[26], line 1
----> 1 em_by_comm_parent = carbon_df.groupby(['commodity_simple','parent_type'])['total_emissions_MtCO2e'].mean()
2 type(em_by_comm_parent)
NameError: name 'carbon_df' is not defined
and look at the index to confirm
em_by_comm_parent.index
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[27], line 1
----> 1 em_by_comm_parent.index
NameError: name 'em_by_comm_parent' is not defined
We can use reset_index to change from the multi-index into a new count from 0 index (and make it a DataFrame in the process)
carbon_df.groupby(['commodity_simple','parent_type'])['total_emissions_MtCO2e'].mean().reset_index()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[28], line 1
----> 1 carbon_df.groupby(['commodity_simple','parent_type'])['total_emissions_MtCO2e'].mean().reset_index()
NameError: name 'carbon_df' is not defined
4.7. Prepare for next class#
Seaborn is a plotting library that gives us opinionated defaults
Important
Run this line one time before class on Thursday
import seaborn as sns
If you get ModuleNotFound error, open a terminal tab and run pip install seaborn .
seaborn’s alias is sns as an inside joke among the developers to the character, Samual Norman Seaborn from West Wing they named the library after per their FAQ
4.8. Questions After Class#
Important
Questions about the assignment have been addreessed by adding hints and tips to the end of the instructions
4.8.1. when putting the two column names ["commodity_simple", "parent_type"] in the square brackets, what did that do?#
We cann answer a question like this by inspecting the code further
First we can look directly at it:
["commodity_simple", "parent_type"]
['commodity_simple', 'parent_type']
Next, we can check its type:
type(["commodity_simple", "parent_type"])
list
It makes a list which is then compatible with the groupby method.
We can look a its help to remember what it can be passed:
# the __doc__ attribute is a property of all functions
# it is a string so I used split to break it into lines,
# looked to see how many i needed then joined them back together
# and printed them (to have the newkines render)
print('\n'.join(carbon_df.groupby.__doc__.split('\n')[:18]))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[32], line 5
1 # the __doc__ attribute is a property of all functions
2 # it is a string so I used split to break it into lines,
3 # looked to see how many i needed then joined them back together
4 # and printed them (to have the newkines render)
----> 5 print('\n'.join(carbon_df.groupby.__doc__.split('\n')[:18]))
NameError: name 'carbon_df' is not defined
4.8.2. How does the apply function work?#
I have explained it and linked resources above, plus the pandas docs
have a section on it in their user guide
4.8.3. How should we be practicing this material. Its hard to tell whats the specific take away from this class vs memorizing methods.#
Review the notes and see the analysis.
What we did is ann example data anlysis. We asked and answered several questions. This serves as example questions and types of questions. How to interpret the statistics.
4.8.4. what does the lambda do inside of the function we created to group the coal commodity#
lambda is the keyword to define a function in line, like def is in a full length definition.
4.8.5. Is there a way to scrape data from a website and make it a csv file if the website does not allow for exporting?#
Sometimes, we will learn webscraping in a few weeks. For now, download locally, or try a different dataset.
4.8.6. Will be working on forms of data visualization?#
Yes