6. Tidy Data and Structural Repairs#
6.1. Intro#
This week, we’ll be cleaning data.
Cleaning data is labor intensive and requires making subjective choices.
We’ll focus on, and assess you on, manipulating data correctly, making reasonable
choices, and documenting the choices you make carefully.
We’ll focus on the programming tools that get used in cleaning data in class this week:
reshaping data
handling missing or incorrect values
renaming columns
Warning
this is incomplete, but will get filled in this week.
import pandas as pd
import seaborn as sns
# make plots look nicer and increase font size
sns.set_theme(font_scale=2, palette='colorblind')
6.2. What is Tidy Data#
Read in the three csv files described below and store them in a list of dataFrames
url_base = 'https://raw.githubusercontent.com/rhodyprog4ds/rhodyds/main/data/'
datasets = ['study_a.csv','study_b.csv','study_c.csv']
study_df_list = [pd.read_csv(url_base +cur_study,na_values='-')
for cur_study in datasets]
study_df_list[0]
| name | treatmenta | treatmentb | |
|---|---|---|---|
| 0 | John Smith | NaN | 2 |
| 1 | Jane Doe | 16.0 | 11 |
| 2 | Mary Johnson | 3.0 | 1 |
study_df_list[1]
| intervention | John Smith | Jane Doe | Mary Johnson | |
|---|---|---|---|---|
| 0 | treatmenta | NaN | 16 | 3 |
| 1 | treatmentb | 2.0 | 11 | 1 |
study_df_list[2]
| person | treatment | result | |
|---|---|---|---|
| 0 | John Smith | a | NaN |
| 1 | Jane Doe | a | 16.0 |
| 2 | Mary Johnson | a | 3.0 |
| 3 | John Smith | b | 2.0 |
| 4 | Jane Doe | b | 11.0 |
| 5 | Mary Johnson | b | 1.0 |
These three all show the same data, but let’s say we have two goals:
find the average effect per person across treatments
find the average effect per treatment across people
This works differently for these three versions.
df_a = study_df_list[0]
df_a.mean(numeric_only=True,)
treatmenta 9.500000
treatmentb 4.666667
dtype: float64
we get the average per treatment, but to get the average per person, we have to go across rows, which we can do here, but doesn’t work as well with plotting
we can work across rows with the axis parameter if needed
df_a.mean(numeric_only=True,axis=1)
0 2.0
1 13.5
2 2.0
dtype: float64
df_b = study_df_list[1]
df_b
| intervention | John Smith | Jane Doe | Mary Johnson | |
|---|---|---|---|---|
| 0 | treatmenta | NaN | 16 | 3 |
| 1 | treatmentb | 2.0 | 11 | 1 |
Now we get the average per person, but what about per treatment? again we have to go across rows instead.
study_df_list[2]
| person | treatment | result | |
|---|---|---|---|
| 0 | John Smith | a | NaN |
| 1 | Jane Doe | a | 16.0 |
| 2 | Mary Johnson | a | 3.0 |
| 3 | John Smith | b | 2.0 |
| 4 | Jane Doe | b | 11.0 |
| 5 | Mary Johnson | b | 1.0 |
For the third one, however, we can use groupby, because this one is tidy.
study_df_list[2].groupby('treatment').mean(numeric_only=True)
| result | |
|---|---|
| treatment | |
| a | 9.500000 |
| b | 4.666667 |
study_df_list[2].groupby('person').mean(numeric_only=True)
| result | |
|---|---|
| person | |
| Jane Doe | 13.5 |
| John Smith | 2.0 |
| Mary Johnson | 2.0 |
The original Tidy Data paper is worth reading to build a deeper understanding of these ideas.
6.3. Tidying Data#
Let’s reshape the first one to match the tidy one. First, we will save it to a DataFrame, this makes things easier to read and enables us to use the built in help in jupyter, because it can’t check types too many levels into a data structure.
df_a.melt(id_vars='name', value_vars=['treatmenta','treatmentb'])
| name | variable | value | |
|---|---|---|---|
| 0 | John Smith | treatmenta | NaN |
| 1 | Jane Doe | treatmenta | 16.0 |
| 2 | Mary Johnson | treatmenta | 3.0 |
| 3 | John Smith | treatmentb | 2.0 |
| 4 | Jane Doe | treatmentb | 11.0 |
| 5 | Mary Johnson | treatmentb | 1.0 |
When we melt a dataset:
the
id_varsstay as columnsthe data from the
value_varscolumns become the values in thevaluecolumnthe column names from the
value_varsbecome the values in thevariablecolumnwe can rename the value and the variable columns.
df_a.melt(id_vars='name', value_vars=['treatmenta','treatmentb'],
var_name= 'treatment_type',
value_name='result',)
| name | treatment_type | result | |
|---|---|---|---|
| 0 | John Smith | treatmenta | NaN |
| 1 | Jane Doe | treatmenta | 16.0 |
| 2 | Mary Johnson | treatmenta | 3.0 |
| 3 | John Smith | treatmentb | 2.0 |
| 4 | Jane Doe | treatmentb | 11.0 |
| 5 | Mary Johnson | treatmentb | 1.0 |
6.4. Transforming the Coffee Data#
Let’s do it for our coffee data:
arabica_data_url = 'https://raw.githubusercontent.com/jldbc/coffee-quality-database/master/data/arabica_data_cleaned.csv'
# load the data
coffee_df = pd.read_csv(arabica_data_url, index_col=0)
coffee_df.head(2)
| Species | Owner | Country.of.Origin | Farm.Name | Lot.Number | Mill | ICO.Number | Company | Altitude | Region | ... | Color | Category.Two.Defects | Expiration | Certification.Body | Certification.Address | Certification.Contact | unit_of_measurement | altitude_low_meters | altitude_high_meters | altitude_mean_meters | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Arabica | metad plc | Ethiopia | metad plc | NaN | metad plc | 2014/2015 | metad agricultural developmet plc | 1950-2200 | guji-hambela | ... | Green | 0 | April 3rd, 2016 | METAD Agricultural Development plc | 309fcf77415a3661ae83e027f7e5f05dad786e44 | 19fef5a731de2db57d16da10287413f5f99bc2dd | m | 1950.0 | 2200.0 | 2075.0 |
| 2 | Arabica | metad plc | Ethiopia | metad plc | NaN | metad plc | 2014/2015 | metad agricultural developmet plc | 1950-2200 | guji-hambela | ... | Green | 1 | April 3rd, 2016 | METAD Agricultural Development plc | 309fcf77415a3661ae83e027f7e5f05dad786e44 | 19fef5a731de2db57d16da10287413f5f99bc2dd | m | 1950.0 | 2200.0 | 2075.0 |
2 rows × 43 columns
coffee_df.columns
Index(['Species', 'Owner', 'Country.of.Origin', 'Farm.Name', 'Lot.Number',
'Mill', 'ICO.Number', 'Company', 'Altitude', 'Region', 'Producer',
'Number.of.Bags', 'Bag.Weight', 'In.Country.Partner', 'Harvest.Year',
'Grading.Date', 'Owner.1', 'Variety', 'Processing.Method', 'Aroma',
'Flavor', 'Aftertaste', 'Acidity', 'Body', 'Balance', 'Uniformity',
'Clean.Cup', 'Sweetness', 'Cupper.Points', 'Total.Cup.Points',
'Moisture', 'Category.One.Defects', 'Quakers', 'Color',
'Category.Two.Defects', 'Expiration', 'Certification.Body',
'Certification.Address', 'Certification.Contact', 'unit_of_measurement',
'altitude_low_meters', 'altitude_high_meters', 'altitude_mean_meters'],
dtype='object')
scores_of_interest = ['Aroma',
'Flavor', 'Aftertaste', 'Acidity', 'Body', 'Balance', 'Uniformity',
'Clean.Cup', 'Sweetness',]
coffee_df[scores_of_interest].head(2)
| Aroma | Flavor | Aftertaste | Acidity | Body | Balance | Uniformity | Clean.Cup | Sweetness | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 8.67 | 8.83 | 8.67 | 8.75 | 8.50 | 8.42 | 10.0 | 10.0 | 10.0 |
| 2 | 8.75 | 8.67 | 8.50 | 8.58 | 8.42 | 8.42 | 10.0 | 10.0 | 10.0 |
We can make this tall using melt to transform with that set of questions and then rename the value and variable columns to be descriptive.
coffee_df_tall = coffee_df.melt(id_vars='Country.of.Origin',value_vars=scores_of_interest,
var_name='rating_type',
value_name='score')
coffee_df_tall.head(1)
| Country.of.Origin | rating_type | score | |
|---|---|---|---|
| 0 | Ethiopia | Aroma | 8.67 |
Notice that the actual column names inside the scores_of_interest variable become the values in the variable column.
This one we can plot in more ways:
sns.displot(data = coffee_df_tall, kind = 'kde', x='score',
col='rating_type', hue='Country.of.Origin',col_wrap=3)
/tmp/ipykernel_2149/2608901824.py:1: UserWarning: Dataset has 0 variance; skipping density estimate. Pass `warn_singular=False` to disable this warning.
sns.displot(data = coffee_df_tall, kind = 'kde', x='score',
<seaborn.axisgrid.FacetGrid at 0x7f23c4c3fc10>
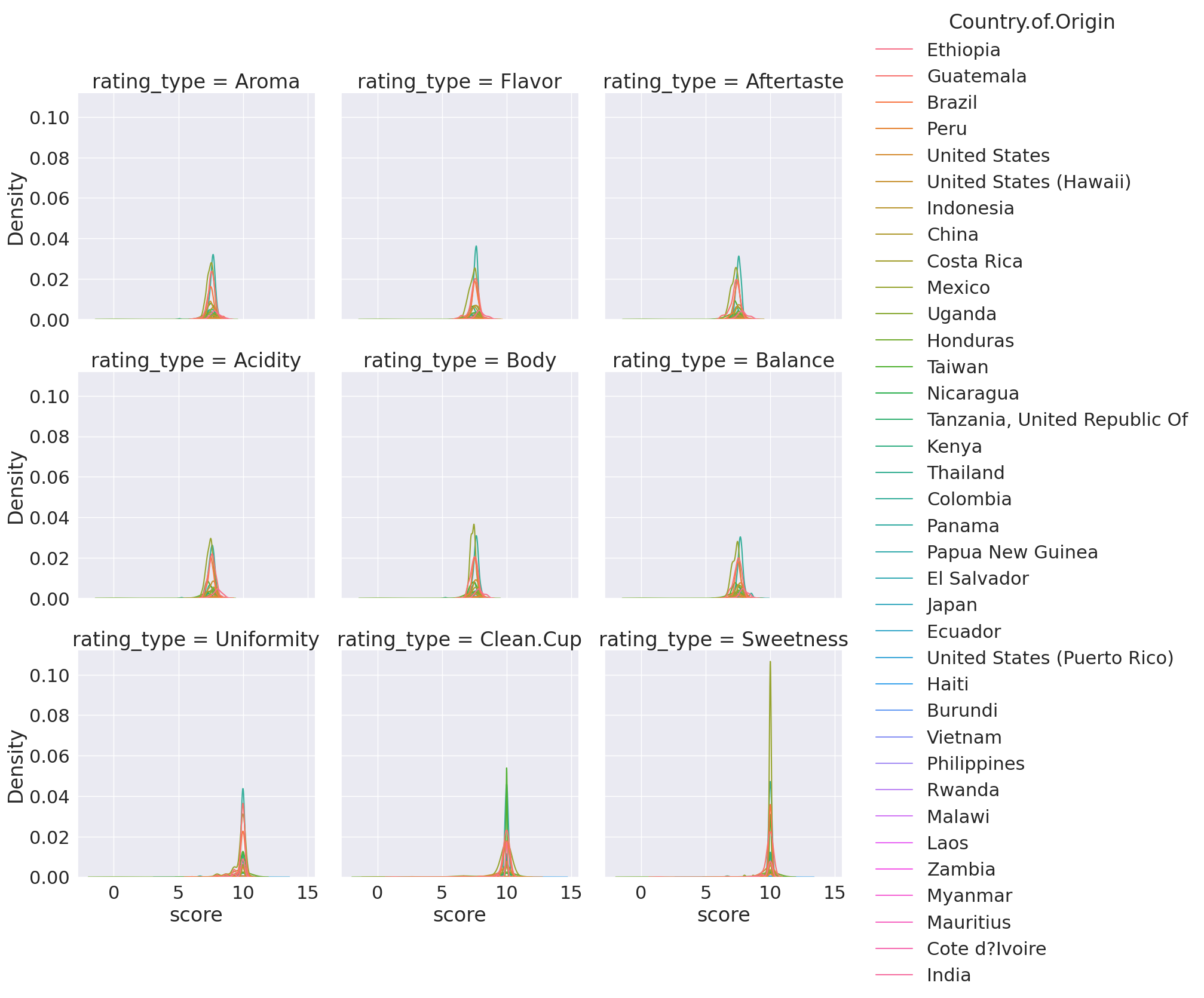
find why warning
6.5. Filtering Data#
Warning
this was not in class this week, but is added here for completeness
high_prod = coffee_df[coffee_df['Number.of.Bags']>250]
high_prod.shape
(368, 43)
coffee_df.shape
(1311, 43)
We see that filters and reduces. We can use any boolean expression in the square brackets.
top_balance = coffee_df[coffee_df['Balance']>coffee_df['Balance'].quantile(.75)]
top_balance.shape
(252, 43)
We can confirm that we got only the top 25% of balance scores:
top_balance.describe()
| Number.of.Bags | Aroma | Flavor | Aftertaste | Acidity | Body | Balance | Uniformity | Clean.Cup | Sweetness | Cupper.Points | Total.Cup.Points | Moisture | Category.One.Defects | Quakers | Category.Two.Defects | altitude_low_meters | altitude_high_meters | altitude_mean_meters | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 252.00000 | 252.000000 | 252.000000 | 252.000000 | 252.000000 | 190.000000 | 190.000000 | 190.000000 |
| mean | 153.337302 | 7.808889 | 7.837659 | 7.734167 | 7.824881 | 7.780159 | 8.003095 | 9.885278 | 9.896667 | 9.885952 | 7.865714 | 84.52504 | 0.072579 | 0.361111 | 0.150794 | 2.801587 | 1343.138168 | 1424.547053 | 1383.842611 |
| std | 126.498576 | 0.355319 | 0.318172 | 0.302481 | 0.320253 | 0.317712 | 0.213056 | 0.363303 | 0.470514 | 0.380433 | 0.383738 | 1.91563 | 0.051682 | 1.249923 | 0.692216 | 4.659771 | 480.927004 | 503.063351 | 484.286247 |
| min | 0.000000 | 5.080000 | 7.170000 | 6.920000 | 7.080000 | 5.250000 | 7.830000 | 6.670000 | 5.330000 | 6.670000 | 5.170000 | 77.00000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 |
| 25% | 12.750000 | 7.647500 | 7.670000 | 7.500000 | 7.670000 | 7.670000 | 7.830000 | 10.000000 | 10.000000 | 10.000000 | 7.670000 | 83.42000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1162.500000 | 1200.000000 | 1200.000000 |
| 50% | 165.500000 | 7.780000 | 7.830000 | 7.750000 | 7.830000 | 7.750000 | 7.920000 | 10.000000 | 10.000000 | 10.000000 | 7.830000 | 84.46000 | 0.100000 | 0.000000 | 0.000000 | 1.000000 | 1400.000000 | 1500.000000 | 1450.000000 |
| 75% | 275.000000 | 8.000000 | 8.000000 | 7.920000 | 8.000000 | 7.920000 | 8.080000 | 10.000000 | 10.000000 | 10.000000 | 8.080000 | 85.52000 | 0.110000 | 0.000000 | 0.000000 | 3.000000 | 1695.000000 | 1800.000000 | 1750.000000 |
| max | 360.000000 | 8.750000 | 8.830000 | 8.670000 | 8.750000 | 8.580000 | 8.750000 | 10.000000 | 10.000000 | 10.000000 | 9.250000 | 90.58000 | 0.210000 | 10.000000 | 6.000000 | 40.000000 | 2560.000000 | 2560.000000 | 2560.000000 |
We can also use the isin method to filter by comparing to an iterable type
total_per_country = coffee_df.groupby('Country.of.Origin')['Number.of.Bags'].sum()
top_countries = total_per_country.sort_values(ascending=False)[:10].index
top_coffee_df = coffee_df[coffee_df['Country.of.Origin'].isin(top_countries)]
6.6. More manipulations#
Warning
this was not in class this week, but is added here for completeness
Here, we will make a tiny DataFrame from scratch to illustrate a couple of points
large_num_df = pd.DataFrame(data= [[730000000,392000000,580200000],
[315040009,580000000,967290000]],
columns = ['a','b','c'])
large_num_df
| a | b | c | |
|---|---|---|---|
| 0 | 730000000 | 392000000 | 580200000 |
| 1 | 315040009 | 580000000 | 967290000 |
This dataet is not tidy, but making it this way was faster to set it up. We could make it tidy using melt as is.
large_num_df.melt()
| variable | value | |
|---|---|---|
| 0 | a | 730000000 |
| 1 | a | 315040009 |
| 2 | b | 392000000 |
| 3 | b | 580000000 |
| 4 | c | 580200000 |
| 5 | c | 967290000 |
However, I want an additional variable, so I wil reset the index, which adds an index column for the original index and adds a new index that is numerical. In this case they’re the same.
large_num_df.reset_index()
| index | a | b | c | |
|---|---|---|---|---|
| 0 | 0 | 730000000 | 392000000 | 580200000 |
| 1 | 1 | 315040009 | 580000000 | 967290000 |
If I melt this one, using the index as the id, then I get a reasonable tidy DataFrame
ls_tall_df = large_num_df.reset_index().melt(id_vars='index')
ls_tall_df
| index | variable | value | |
|---|---|---|---|
| 0 | 0 | a | 730000000 |
| 1 | 1 | a | 315040009 |
| 2 | 0 | b | 392000000 |
| 3 | 1 | b | 580000000 |
| 4 | 0 | c | 580200000 |
| 5 | 1 | c | 967290000 |
Now, we can plot.
sns.catplot(data = ls_tall_df,x='variable',y='value',
hue='index',kind='bar')
<seaborn.axisgrid.FacetGrid at 0x7f23bf8dde80>
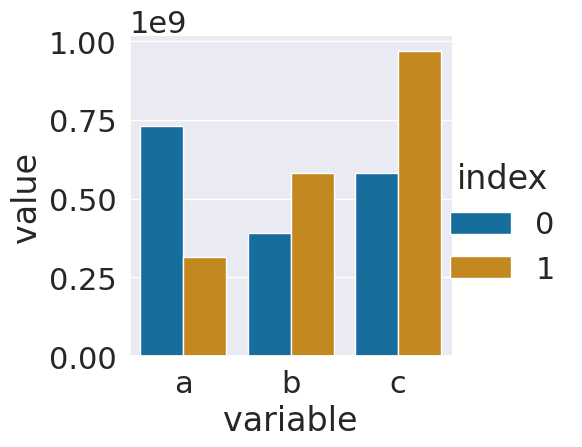
Since the numbers are so big, this might be hard to interpret. Displaying it with all the 0s would not be easier to read. The best thing to do is to add a new colum with adjusted values and a corresponding title.
ls_tall_df['value (millions)'] = ls_tall_df['value']/1000000
ls_tall_df.head()
| index | variable | value | value (millions) | |
|---|---|---|---|---|
| 0 | 0 | a | 730000000 | 730.000000 |
| 1 | 1 | a | 315040009 | 315.040009 |
| 2 | 0 | b | 392000000 | 392.000000 |
| 3 | 1 | b | 580000000 | 580.000000 |
| 4 | 0 | c | 580200000 | 580.200000 |
Now we can plot again, with the smaller values and an updated axis label. Adding a column with the adjusted title is good practice because it does not lose any data and since we set the value and the title at the same time it keeps it clear what the values are.
6.7. Questions after Class#
6.7.1. In the data we had about treatment a and treatment b. Say there was another column that provided information about the age of the people. Could we create another value variable to or would we put it in the value_vars list along with treatment a and treatmentb?#
We can have multiple variables as the id_vars so that the dataset would have 4 columns instead of 3.
6.7.2. Are there any prebuilt methods to identify and remove extreme outliers?#
There may be, that is a good thing to look in the documentation for. However, typically the definition of an outlier is best made within context, so the filtering strategy that we just used would be the way to remove them, after doign some EDA.
6.7.3. Is there a specific metric to which a dataset must meet to consider it ‘cleaned’?#
It’s “clean” when it will work for the analysis you want to do. This means clean enough for one analysis may not be enough for another goal. Domain expertise is alwasy important.
6.7.4. how does melt work underneath the hood?#
The melt documentation is the place to read about the details. It also includes alink to the source code if reading how it is implemented in code is more helpful to you can reading English.
6.7.5. Do people ever write their melted datasets to new files for redistribution, maybe to make things easier for future researchers?#
Yes! If you look in the R Tidy Tuesday datsets most distribute the “raw” and cleaned data.
6.7.6. the ending graphs are hard to read. Is there a way to label specific countries kde lines?#
Yes
6.7.7. how do you get the index of something in a row#
I do not understand this question, please reach out with more info so that I can help you
6.7.8. i was trying to do somthing earlier where I got the max of a column but wanted to know somthing else associated with that row#
most pandas methods allow you to use axis to apply them across rows instead of columns
6.7.9. cleaning out different data types#
Once the data is loaded into pandas, we can use all of the same techniques
6.7.10. Is there a scenario where a dataset cannot be cleaned using pandas methods?#
yes, there are cases where cleaning the already loaded dataframe is not the best way to make the necessary fixes. One tool that can help is called Openrefine it is an open source program that provides a GUI based interface to clean data, but also outputs a script of what actions were taking, to make the cleaning replicable.
This is something you could learn about and try for the level 3 prepare achievement in your portfolio. One resource for learning it is the Data Carpentry lesson on Data Cleaning for Ecologists
6.7.11. Is there another standard format for sharing cleaned datasets?#
.csv is the widely accepted preferred format for well strucutured tabular data that is of reasonable size. If it is too large, a database might (next week!) can be useful.
6.7.12. With extremely large datasets, is there a way to find what pieces of the dataset are missing values other than manually checking#
Using strategies like we did to day, by testing dropping and the EDA strategies that we used last week can help. If it is a very large dataset, you may need to use more powerful system, but the basics work out the same.
Also, remember in most contexts, you will have some relevant domain knowledge that can help you.