7. Tidy Data and Reshaping Datasets#
import pandas as pd
import seaborn as sns
sns.set_theme(palette='colorblind',font_scale=2)
url_base = 'https://raw.githubusercontent.com/rhodyprog4ds/rhodyds/main/data/'
datasets = ['study_a.csv','study_b.csv','study_c.csv']
list_of_df = [pd.read_csv(url_base + dataset,na_values='-') for dataset in datasets]
list_of_df[0]
| name | treatmenta | treatmentb | |
|---|---|---|---|
| 0 | John Smith | NaN | 2 |
| 1 | Jane Doe | 16.0 | 11 |
| 2 | Mary Johnson | 3.0 | 1 |
list_of_df[1]
| intervention | John Smith | Jane Doe | Mary Johnson | |
|---|---|---|---|---|
| 0 | treatmenta | NaN | 16 | 3 |
| 1 | treatmentb | 2.0 | 11 | 1 |
list_of_df[2]
| person | treatment | result | |
|---|---|---|---|
| 0 | John Smith | a | NaN |
| 1 | Jane Doe | a | 16.0 |
| 2 | Mary Johnson | a | 3.0 |
| 3 | John Smith | b | 2.0 |
| 4 | Jane Doe | b | 11.0 |
| 5 | Mary Johnson | b | 1.0 |
list_of_df[2].mean()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1680, in _ensure_numeric(x)
1679 try:
-> 1680 x = x.astype(np.complex128)
1681 except (TypeError, ValueError):
ValueError: complex() arg is a malformed string
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1683, in _ensure_numeric(x)
1682 try:
-> 1683 x = x.astype(np.float64)
1684 except ValueError as err:
1685 # GH#29941 we get here with object arrays containing strs
ValueError: could not convert string to float: 'John SmithJane DoeMary JohnsonJohn SmithJane DoeMary Johnson'
The above exception was the direct cause of the following exception:
TypeError Traceback (most recent call last)
Cell In[7], line 1
----> 1 list_of_df[2].mean()
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11563, in NDFrame._add_numeric_operations.<locals>.mean(self, axis, skipna, numeric_only, **kwargs)
11546 @doc(
11547 _num_doc,
11548 desc="Return the mean of the values over the requested axis.",
(...)
11561 **kwargs,
11562 ):
> 11563 return NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11208, in NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
11201 def mean(
11202 self,
11203 axis: Axis | None = 0,
(...)
11206 **kwargs,
11207 ) -> Series | float:
> 11208 return self._stat_function(
11209 "mean", nanops.nanmean, axis, skipna, numeric_only, **kwargs
11210 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11165, in NDFrame._stat_function(self, name, func, axis, skipna, numeric_only, **kwargs)
11161 nv.validate_stat_func((), kwargs, fname=name)
11163 validate_bool_kwarg(skipna, "skipna", none_allowed=False)
> 11165 return self._reduce(
11166 func, name=name, axis=axis, skipna=skipna, numeric_only=numeric_only
11167 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/frame.py:10519, in DataFrame._reduce(self, op, name, axis, skipna, numeric_only, filter_type, **kwds)
10515 df = df.T
10517 # After possibly _get_data and transposing, we are now in the
10518 # simple case where we can use BlockManager.reduce
> 10519 res = df._mgr.reduce(blk_func)
10520 out = df._constructor(res).iloc[0]
10521 if out_dtype is not None:
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/internals/managers.py:1534, in BlockManager.reduce(self, func)
1532 res_blocks: list[Block] = []
1533 for blk in self.blocks:
-> 1534 nbs = blk.reduce(func)
1535 res_blocks.extend(nbs)
1537 index = Index([None]) # placeholder
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/internals/blocks.py:339, in Block.reduce(self, func)
333 @final
334 def reduce(self, func) -> list[Block]:
335 # We will apply the function and reshape the result into a single-row
336 # Block with the same mgr_locs; squeezing will be done at a higher level
337 assert self.ndim == 2
--> 339 result = func(self.values)
341 if self.values.ndim == 1:
342 # TODO(EA2D): special case not needed with 2D EAs
343 res_values = np.array([[result]])
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/frame.py:10482, in DataFrame._reduce.<locals>.blk_func(values, axis)
10480 return values._reduce(name, skipna=skipna, **kwds)
10481 else:
> 10482 return op(values, axis=axis, skipna=skipna, **kwds)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:96, in disallow.__call__.<locals>._f(*args, **kwargs)
94 try:
95 with np.errstate(invalid="ignore"):
---> 96 return f(*args, **kwargs)
97 except ValueError as e:
98 # we want to transform an object array
99 # ValueError message to the more typical TypeError
100 # e.g. this is normally a disallowed function on
101 # object arrays that contain strings
102 if is_object_dtype(args[0]):
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:158, in bottleneck_switch.__call__.<locals>.f(values, axis, skipna, **kwds)
156 result = alt(values, axis=axis, skipna=skipna, **kwds)
157 else:
--> 158 result = alt(values, axis=axis, skipna=skipna, **kwds)
160 return result
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:421, in _datetimelike_compat.<locals>.new_func(values, axis, skipna, mask, **kwargs)
418 if datetimelike and mask is None:
419 mask = isna(values)
--> 421 result = func(values, axis=axis, skipna=skipna, mask=mask, **kwargs)
423 if datetimelike:
424 result = _wrap_results(result, orig_values.dtype, fill_value=iNaT)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:727, in nanmean(values, axis, skipna, mask)
724 dtype_count = dtype
726 count = _get_counts(values.shape, mask, axis, dtype=dtype_count)
--> 727 the_sum = _ensure_numeric(values.sum(axis, dtype=dtype_sum))
729 if axis is not None and getattr(the_sum, "ndim", False):
730 count = cast(np.ndarray, count)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1686, in _ensure_numeric(x)
1683 x = x.astype(np.float64)
1684 except ValueError as err:
1685 # GH#29941 we get here with object arrays containing strs
-> 1686 raise TypeError(f"Could not convert {x} to numeric") from err
1687 else:
1688 if not np.any(np.imag(x)):
TypeError: Could not convert ['John SmithJane DoeMary JohnsonJohn SmithJane DoeMary Johnson' 'aaabbb'] to numeric
sum([16,3,2,11,1])/5
6.6
sum([16,3,2,11,1,0])/6
5.5
list_of_df[2].groupby('treatment').mean()
---------------------------------------------------------------------------
NotImplementedError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1490, in GroupBy._cython_agg_general.<locals>.array_func(values)
1489 try:
-> 1490 result = self.grouper._cython_operation(
1491 "aggregate",
1492 values,
1493 how,
1494 axis=data.ndim - 1,
1495 min_count=min_count,
1496 **kwargs,
1497 )
1498 except NotImplementedError:
1499 # generally if we have numeric_only=False
1500 # and non-applicable functions
1501 # try to python agg
1502 # TODO: shouldn't min_count matter?
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:959, in BaseGrouper._cython_operation(self, kind, values, how, axis, min_count, **kwargs)
958 ngroups = self.ngroups
--> 959 return cy_op.cython_operation(
960 values=values,
961 axis=axis,
962 min_count=min_count,
963 comp_ids=ids,
964 ngroups=ngroups,
965 **kwargs,
966 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:657, in WrappedCythonOp.cython_operation(self, values, axis, min_count, comp_ids, ngroups, **kwargs)
649 return self._ea_wrap_cython_operation(
650 values,
651 min_count=min_count,
(...)
654 **kwargs,
655 )
--> 657 return self._cython_op_ndim_compat(
658 values,
659 min_count=min_count,
660 ngroups=ngroups,
661 comp_ids=comp_ids,
662 mask=None,
663 **kwargs,
664 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:497, in WrappedCythonOp._cython_op_ndim_compat(self, values, min_count, ngroups, comp_ids, mask, result_mask, **kwargs)
495 return res.T
--> 497 return self._call_cython_op(
498 values,
499 min_count=min_count,
500 ngroups=ngroups,
501 comp_ids=comp_ids,
502 mask=mask,
503 result_mask=result_mask,
504 **kwargs,
505 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:541, in WrappedCythonOp._call_cython_op(self, values, min_count, ngroups, comp_ids, mask, result_mask, **kwargs)
540 out_shape = self._get_output_shape(ngroups, values)
--> 541 func = self._get_cython_function(self.kind, self.how, values.dtype, is_numeric)
542 values = self._get_cython_vals(values)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:173, in WrappedCythonOp._get_cython_function(cls, kind, how, dtype, is_numeric)
171 if "object" not in f.__signatures__:
172 # raise NotImplementedError here rather than TypeError later
--> 173 raise NotImplementedError(
174 f"function is not implemented for this dtype: "
175 f"[how->{how},dtype->{dtype_str}]"
176 )
177 return f
NotImplementedError: function is not implemented for this dtype: [how->mean,dtype->object]
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1692, in _ensure_numeric(x)
1691 try:
-> 1692 x = float(x)
1693 except (TypeError, ValueError):
1694 # e.g. "1+1j" or "foo"
ValueError: could not convert string to float: 'John SmithJane DoeMary Johnson'
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1696, in _ensure_numeric(x)
1695 try:
-> 1696 x = complex(x)
1697 except ValueError as err:
1698 # e.g. "foo"
ValueError: complex() arg is a malformed string
The above exception was the direct cause of the following exception:
TypeError Traceback (most recent call last)
Cell In[10], line 1
----> 1 list_of_df[2].groupby('treatment').mean()
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1855, in GroupBy.mean(self, numeric_only, engine, engine_kwargs)
1853 return self._numba_agg_general(sliding_mean, engine_kwargs)
1854 else:
-> 1855 result = self._cython_agg_general(
1856 "mean",
1857 alt=lambda x: Series(x).mean(numeric_only=numeric_only),
1858 numeric_only=numeric_only,
1859 )
1860 return result.__finalize__(self.obj, method="groupby")
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1507, in GroupBy._cython_agg_general(self, how, alt, numeric_only, min_count, **kwargs)
1503 result = self._agg_py_fallback(values, ndim=data.ndim, alt=alt)
1505 return result
-> 1507 new_mgr = data.grouped_reduce(array_func)
1508 res = self._wrap_agged_manager(new_mgr)
1509 out = self._wrap_aggregated_output(res)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/internals/managers.py:1503, in BlockManager.grouped_reduce(self, func)
1499 if blk.is_object:
1500 # split on object-dtype blocks bc some columns may raise
1501 # while others do not.
1502 for sb in blk._split():
-> 1503 applied = sb.apply(func)
1504 result_blocks = extend_blocks(applied, result_blocks)
1505 else:
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/internals/blocks.py:329, in Block.apply(self, func, **kwargs)
323 @final
324 def apply(self, func, **kwargs) -> list[Block]:
325 """
326 apply the function to my values; return a block if we are not
327 one
328 """
--> 329 result = func(self.values, **kwargs)
331 return self._split_op_result(result)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1503, in GroupBy._cython_agg_general.<locals>.array_func(values)
1490 result = self.grouper._cython_operation(
1491 "aggregate",
1492 values,
(...)
1496 **kwargs,
1497 )
1498 except NotImplementedError:
1499 # generally if we have numeric_only=False
1500 # and non-applicable functions
1501 # try to python agg
1502 # TODO: shouldn't min_count matter?
-> 1503 result = self._agg_py_fallback(values, ndim=data.ndim, alt=alt)
1505 return result
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1457, in GroupBy._agg_py_fallback(self, values, ndim, alt)
1452 ser = df.iloc[:, 0]
1454 # We do not get here with UDFs, so we know that our dtype
1455 # should always be preserved by the implemented aggregations
1456 # TODO: Is this exactly right; see WrappedCythonOp get_result_dtype?
-> 1457 res_values = self.grouper.agg_series(ser, alt, preserve_dtype=True)
1459 if isinstance(values, Categorical):
1460 # Because we only get here with known dtype-preserving
1461 # reductions, we cast back to Categorical.
1462 # TODO: if we ever get "rank" working, exclude it here.
1463 res_values = type(values)._from_sequence(res_values, dtype=values.dtype)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:994, in BaseGrouper.agg_series(self, obj, func, preserve_dtype)
987 if len(obj) > 0 and not isinstance(obj._values, np.ndarray):
988 # we can preserve a little bit more aggressively with EA dtype
989 # because maybe_cast_pointwise_result will do a try/except
990 # with _from_sequence. NB we are assuming here that _from_sequence
991 # is sufficiently strict that it casts appropriately.
992 preserve_dtype = True
--> 994 result = self._aggregate_series_pure_python(obj, func)
996 npvalues = lib.maybe_convert_objects(result, try_float=False)
997 if preserve_dtype:
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:1015, in BaseGrouper._aggregate_series_pure_python(self, obj, func)
1012 splitter = self._get_splitter(obj, axis=0)
1014 for i, group in enumerate(splitter):
-> 1015 res = func(group)
1016 res = libreduction.extract_result(res)
1018 if not initialized:
1019 # We only do this validation on the first iteration
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1857, in GroupBy.mean.<locals>.<lambda>(x)
1853 return self._numba_agg_general(sliding_mean, engine_kwargs)
1854 else:
1855 result = self._cython_agg_general(
1856 "mean",
-> 1857 alt=lambda x: Series(x).mean(numeric_only=numeric_only),
1858 numeric_only=numeric_only,
1859 )
1860 return result.__finalize__(self.obj, method="groupby")
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11563, in NDFrame._add_numeric_operations.<locals>.mean(self, axis, skipna, numeric_only, **kwargs)
11546 @doc(
11547 _num_doc,
11548 desc="Return the mean of the values over the requested axis.",
(...)
11561 **kwargs,
11562 ):
> 11563 return NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11208, in NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
11201 def mean(
11202 self,
11203 axis: Axis | None = 0,
(...)
11206 **kwargs,
11207 ) -> Series | float:
> 11208 return self._stat_function(
11209 "mean", nanops.nanmean, axis, skipna, numeric_only, **kwargs
11210 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11165, in NDFrame._stat_function(self, name, func, axis, skipna, numeric_only, **kwargs)
11161 nv.validate_stat_func((), kwargs, fname=name)
11163 validate_bool_kwarg(skipna, "skipna", none_allowed=False)
> 11165 return self._reduce(
11166 func, name=name, axis=axis, skipna=skipna, numeric_only=numeric_only
11167 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/series.py:4671, in Series._reduce(self, op, name, axis, skipna, numeric_only, filter_type, **kwds)
4666 raise TypeError(
4667 f"Series.{name} does not allow {kwd_name}={numeric_only} "
4668 "with non-numeric dtypes."
4669 )
4670 with np.errstate(all="ignore"):
-> 4671 return op(delegate, skipna=skipna, **kwds)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:96, in disallow.__call__.<locals>._f(*args, **kwargs)
94 try:
95 with np.errstate(invalid="ignore"):
---> 96 return f(*args, **kwargs)
97 except ValueError as e:
98 # we want to transform an object array
99 # ValueError message to the more typical TypeError
100 # e.g. this is normally a disallowed function on
101 # object arrays that contain strings
102 if is_object_dtype(args[0]):
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:158, in bottleneck_switch.__call__.<locals>.f(values, axis, skipna, **kwds)
156 result = alt(values, axis=axis, skipna=skipna, **kwds)
157 else:
--> 158 result = alt(values, axis=axis, skipna=skipna, **kwds)
160 return result
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:421, in _datetimelike_compat.<locals>.new_func(values, axis, skipna, mask, **kwargs)
418 if datetimelike and mask is None:
419 mask = isna(values)
--> 421 result = func(values, axis=axis, skipna=skipna, mask=mask, **kwargs)
423 if datetimelike:
424 result = _wrap_results(result, orig_values.dtype, fill_value=iNaT)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:727, in nanmean(values, axis, skipna, mask)
724 dtype_count = dtype
726 count = _get_counts(values.shape, mask, axis, dtype=dtype_count)
--> 727 the_sum = _ensure_numeric(values.sum(axis, dtype=dtype_sum))
729 if axis is not None and getattr(the_sum, "ndim", False):
730 count = cast(np.ndarray, count)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1699, in _ensure_numeric(x)
1696 x = complex(x)
1697 except ValueError as err:
1698 # e.g. "foo"
-> 1699 raise TypeError(f"Could not convert {x} to numeric") from err
1700 return x
TypeError: Could not convert John SmithJane DoeMary Johnson to numeric
list_of_df[2].groupby('person').mean()
---------------------------------------------------------------------------
NotImplementedError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1490, in GroupBy._cython_agg_general.<locals>.array_func(values)
1489 try:
-> 1490 result = self.grouper._cython_operation(
1491 "aggregate",
1492 values,
1493 how,
1494 axis=data.ndim - 1,
1495 min_count=min_count,
1496 **kwargs,
1497 )
1498 except NotImplementedError:
1499 # generally if we have numeric_only=False
1500 # and non-applicable functions
1501 # try to python agg
1502 # TODO: shouldn't min_count matter?
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:959, in BaseGrouper._cython_operation(self, kind, values, how, axis, min_count, **kwargs)
958 ngroups = self.ngroups
--> 959 return cy_op.cython_operation(
960 values=values,
961 axis=axis,
962 min_count=min_count,
963 comp_ids=ids,
964 ngroups=ngroups,
965 **kwargs,
966 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:657, in WrappedCythonOp.cython_operation(self, values, axis, min_count, comp_ids, ngroups, **kwargs)
649 return self._ea_wrap_cython_operation(
650 values,
651 min_count=min_count,
(...)
654 **kwargs,
655 )
--> 657 return self._cython_op_ndim_compat(
658 values,
659 min_count=min_count,
660 ngroups=ngroups,
661 comp_ids=comp_ids,
662 mask=None,
663 **kwargs,
664 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:497, in WrappedCythonOp._cython_op_ndim_compat(self, values, min_count, ngroups, comp_ids, mask, result_mask, **kwargs)
495 return res.T
--> 497 return self._call_cython_op(
498 values,
499 min_count=min_count,
500 ngroups=ngroups,
501 comp_ids=comp_ids,
502 mask=mask,
503 result_mask=result_mask,
504 **kwargs,
505 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:541, in WrappedCythonOp._call_cython_op(self, values, min_count, ngroups, comp_ids, mask, result_mask, **kwargs)
540 out_shape = self._get_output_shape(ngroups, values)
--> 541 func = self._get_cython_function(self.kind, self.how, values.dtype, is_numeric)
542 values = self._get_cython_vals(values)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:173, in WrappedCythonOp._get_cython_function(cls, kind, how, dtype, is_numeric)
171 if "object" not in f.__signatures__:
172 # raise NotImplementedError here rather than TypeError later
--> 173 raise NotImplementedError(
174 f"function is not implemented for this dtype: "
175 f"[how->{how},dtype->{dtype_str}]"
176 )
177 return f
NotImplementedError: function is not implemented for this dtype: [how->mean,dtype->object]
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1692, in _ensure_numeric(x)
1691 try:
-> 1692 x = float(x)
1693 except (TypeError, ValueError):
1694 # e.g. "1+1j" or "foo"
ValueError: could not convert string to float: 'ab'
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1696, in _ensure_numeric(x)
1695 try:
-> 1696 x = complex(x)
1697 except ValueError as err:
1698 # e.g. "foo"
ValueError: complex() arg is a malformed string
The above exception was the direct cause of the following exception:
TypeError Traceback (most recent call last)
Cell In[11], line 1
----> 1 list_of_df[2].groupby('person').mean()
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1855, in GroupBy.mean(self, numeric_only, engine, engine_kwargs)
1853 return self._numba_agg_general(sliding_mean, engine_kwargs)
1854 else:
-> 1855 result = self._cython_agg_general(
1856 "mean",
1857 alt=lambda x: Series(x).mean(numeric_only=numeric_only),
1858 numeric_only=numeric_only,
1859 )
1860 return result.__finalize__(self.obj, method="groupby")
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1507, in GroupBy._cython_agg_general(self, how, alt, numeric_only, min_count, **kwargs)
1503 result = self._agg_py_fallback(values, ndim=data.ndim, alt=alt)
1505 return result
-> 1507 new_mgr = data.grouped_reduce(array_func)
1508 res = self._wrap_agged_manager(new_mgr)
1509 out = self._wrap_aggregated_output(res)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/internals/managers.py:1503, in BlockManager.grouped_reduce(self, func)
1499 if blk.is_object:
1500 # split on object-dtype blocks bc some columns may raise
1501 # while others do not.
1502 for sb in blk._split():
-> 1503 applied = sb.apply(func)
1504 result_blocks = extend_blocks(applied, result_blocks)
1505 else:
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/internals/blocks.py:329, in Block.apply(self, func, **kwargs)
323 @final
324 def apply(self, func, **kwargs) -> list[Block]:
325 """
326 apply the function to my values; return a block if we are not
327 one
328 """
--> 329 result = func(self.values, **kwargs)
331 return self._split_op_result(result)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1503, in GroupBy._cython_agg_general.<locals>.array_func(values)
1490 result = self.grouper._cython_operation(
1491 "aggregate",
1492 values,
(...)
1496 **kwargs,
1497 )
1498 except NotImplementedError:
1499 # generally if we have numeric_only=False
1500 # and non-applicable functions
1501 # try to python agg
1502 # TODO: shouldn't min_count matter?
-> 1503 result = self._agg_py_fallback(values, ndim=data.ndim, alt=alt)
1505 return result
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1457, in GroupBy._agg_py_fallback(self, values, ndim, alt)
1452 ser = df.iloc[:, 0]
1454 # We do not get here with UDFs, so we know that our dtype
1455 # should always be preserved by the implemented aggregations
1456 # TODO: Is this exactly right; see WrappedCythonOp get_result_dtype?
-> 1457 res_values = self.grouper.agg_series(ser, alt, preserve_dtype=True)
1459 if isinstance(values, Categorical):
1460 # Because we only get here with known dtype-preserving
1461 # reductions, we cast back to Categorical.
1462 # TODO: if we ever get "rank" working, exclude it here.
1463 res_values = type(values)._from_sequence(res_values, dtype=values.dtype)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:994, in BaseGrouper.agg_series(self, obj, func, preserve_dtype)
987 if len(obj) > 0 and not isinstance(obj._values, np.ndarray):
988 # we can preserve a little bit more aggressively with EA dtype
989 # because maybe_cast_pointwise_result will do a try/except
990 # with _from_sequence. NB we are assuming here that _from_sequence
991 # is sufficiently strict that it casts appropriately.
992 preserve_dtype = True
--> 994 result = self._aggregate_series_pure_python(obj, func)
996 npvalues = lib.maybe_convert_objects(result, try_float=False)
997 if preserve_dtype:
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/ops.py:1015, in BaseGrouper._aggregate_series_pure_python(self, obj, func)
1012 splitter = self._get_splitter(obj, axis=0)
1014 for i, group in enumerate(splitter):
-> 1015 res = func(group)
1016 res = libreduction.extract_result(res)
1018 if not initialized:
1019 # We only do this validation on the first iteration
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/groupby/groupby.py:1857, in GroupBy.mean.<locals>.<lambda>(x)
1853 return self._numba_agg_general(sliding_mean, engine_kwargs)
1854 else:
1855 result = self._cython_agg_general(
1856 "mean",
-> 1857 alt=lambda x: Series(x).mean(numeric_only=numeric_only),
1858 numeric_only=numeric_only,
1859 )
1860 return result.__finalize__(self.obj, method="groupby")
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11563, in NDFrame._add_numeric_operations.<locals>.mean(self, axis, skipna, numeric_only, **kwargs)
11546 @doc(
11547 _num_doc,
11548 desc="Return the mean of the values over the requested axis.",
(...)
11561 **kwargs,
11562 ):
> 11563 return NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11208, in NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
11201 def mean(
11202 self,
11203 axis: Axis | None = 0,
(...)
11206 **kwargs,
11207 ) -> Series | float:
> 11208 return self._stat_function(
11209 "mean", nanops.nanmean, axis, skipna, numeric_only, **kwargs
11210 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/generic.py:11165, in NDFrame._stat_function(self, name, func, axis, skipna, numeric_only, **kwargs)
11161 nv.validate_stat_func((), kwargs, fname=name)
11163 validate_bool_kwarg(skipna, "skipna", none_allowed=False)
> 11165 return self._reduce(
11166 func, name=name, axis=axis, skipna=skipna, numeric_only=numeric_only
11167 )
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/series.py:4671, in Series._reduce(self, op, name, axis, skipna, numeric_only, filter_type, **kwds)
4666 raise TypeError(
4667 f"Series.{name} does not allow {kwd_name}={numeric_only} "
4668 "with non-numeric dtypes."
4669 )
4670 with np.errstate(all="ignore"):
-> 4671 return op(delegate, skipna=skipna, **kwds)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:96, in disallow.__call__.<locals>._f(*args, **kwargs)
94 try:
95 with np.errstate(invalid="ignore"):
---> 96 return f(*args, **kwargs)
97 except ValueError as e:
98 # we want to transform an object array
99 # ValueError message to the more typical TypeError
100 # e.g. this is normally a disallowed function on
101 # object arrays that contain strings
102 if is_object_dtype(args[0]):
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:158, in bottleneck_switch.__call__.<locals>.f(values, axis, skipna, **kwds)
156 result = alt(values, axis=axis, skipna=skipna, **kwds)
157 else:
--> 158 result = alt(values, axis=axis, skipna=skipna, **kwds)
160 return result
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:421, in _datetimelike_compat.<locals>.new_func(values, axis, skipna, mask, **kwargs)
418 if datetimelike and mask is None:
419 mask = isna(values)
--> 421 result = func(values, axis=axis, skipna=skipna, mask=mask, **kwargs)
423 if datetimelike:
424 result = _wrap_results(result, orig_values.dtype, fill_value=iNaT)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:727, in nanmean(values, axis, skipna, mask)
724 dtype_count = dtype
726 count = _get_counts(values.shape, mask, axis, dtype=dtype_count)
--> 727 the_sum = _ensure_numeric(values.sum(axis, dtype=dtype_sum))
729 if axis is not None and getattr(the_sum, "ndim", False):
730 count = cast(np.ndarray, count)
File /opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/pandas/core/nanops.py:1699, in _ensure_numeric(x)
1696 x = complex(x)
1697 except ValueError as err:
1698 # e.g. "foo"
-> 1699 raise TypeError(f"Could not convert {x} to numeric") from err
1700 return x
TypeError: Could not convert ab to numeric
dfa = list_of_df[0]
dfa
| name | treatmenta | treatmentb | |
|---|---|---|---|
| 0 | John Smith | NaN | 2 |
| 1 | Jane Doe | 16.0 | 11 |
| 2 | Mary Johnson | 3.0 | 1 |
dfa.melt(id_vars=['name'],var_name='treatment',value_name='result')
| name | treatment | result | |
|---|---|---|---|
| 0 | John Smith | treatmenta | NaN |
| 1 | Jane Doe | treatmenta | 16.0 |
| 2 | Mary Johnson | treatmenta | 3.0 |
| 3 | John Smith | treatmentb | 2.0 |
| 4 | Jane Doe | treatmentb | 11.0 |
| 5 | Mary Johnson | treatmentb | 1.0 |
arabica_data_url = 'https://raw.githubusercontent.com/jldbc/coffee-quality-database/master/data/arabica_data_cleaned.csv'
# load the data
coffee_df = pd.read_csv(arabica_data_url)
# get total bags per country
bags_per_country = coffee_df.groupby('Country.of.Origin')['Number.of.Bags'].sum()
# sort descending, keep only the top 10 and pick out only the country names
top_bags_country_list = bags_per_country.sort_values(ascending=False)[:10].index
# filter the original data for only the countries in the top list
top_coffee_df = coffee_df[coffee_df['Country.of.Origin'].isin(top_bags_country_list)]
bags_per_country
Country.of.Origin
Brazil 30534
Burundi 520
China 55
Colombia 41204
Costa Rica 10354
Cote d?Ivoire 2
Ecuador 1
El Salvador 4449
Ethiopia 11761
Guatemala 36868
Haiti 390
Honduras 13167
India 20
Indonesia 1658
Japan 20
Kenya 3971
Laos 81
Malawi 557
Mauritius 1
Mexico 24140
Myanmar 10
Nicaragua 6406
Panama 537
Papua New Guinea 7
Peru 2336
Philippines 259
Rwanda 150
Taiwan 1914
Tanzania, United Republic Of 3760
Thailand 1310
Uganda 3868
United States 361
United States (Hawaii) 833
United States (Puerto Rico) 71
Vietnam 10
Zambia 13
Name: Number.of.Bags, dtype: int64
top_bags_country_list
Index(['Colombia', 'Guatemala', 'Brazil', 'Mexico', 'Honduras', 'Ethiopia',
'Costa Rica', 'Nicaragua', 'El Salvador', 'Kenya'],
dtype='object', name='Country.of.Origin')
top_coffee_df.head(1)
| Unnamed: 0 | Species | Owner | Country.of.Origin | Farm.Name | Lot.Number | Mill | ICO.Number | Company | Altitude | ... | Color | Category.Two.Defects | Expiration | Certification.Body | Certification.Address | Certification.Contact | unit_of_measurement | altitude_low_meters | altitude_high_meters | altitude_mean_meters | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | Arabica | metad plc | Ethiopia | metad plc | NaN | metad plc | 2014/2015 | metad agricultural developmet plc | 1950-2200 | ... | Green | 0 | April 3rd, 2016 | METAD Agricultural Development plc | 309fcf77415a3661ae83e027f7e5f05dad786e44 | 19fef5a731de2db57d16da10287413f5f99bc2dd | m | 1950.0 | 2200.0 | 2075.0 |
1 rows × 44 columns
coffee_df.head(1)
| Unnamed: 0 | Species | Owner | Country.of.Origin | Farm.Name | Lot.Number | Mill | ICO.Number | Company | Altitude | ... | Color | Category.Two.Defects | Expiration | Certification.Body | Certification.Address | Certification.Contact | unit_of_measurement | altitude_low_meters | altitude_high_meters | altitude_mean_meters | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | Arabica | metad plc | Ethiopia | metad plc | NaN | metad plc | 2014/2015 | metad agricultural developmet plc | 1950-2200 | ... | Green | 0 | April 3rd, 2016 | METAD Agricultural Development plc | 309fcf77415a3661ae83e027f7e5f05dad786e44 | 19fef5a731de2db57d16da10287413f5f99bc2dd | m | 1950.0 | 2200.0 | 2075.0 |
1 rows × 44 columns
coffee_df.shape,top_coffee_df.shape
((1311, 44), (952, 44))
top_coffee_df.describe()
| Unnamed: 0 | Number.of.Bags | Aroma | Flavor | Aftertaste | Acidity | Body | Balance | Uniformity | Clean.Cup | Sweetness | Cupper.Points | Total.Cup.Points | Moisture | Category.One.Defects | Quakers | Category.Two.Defects | altitude_low_meters | altitude_high_meters | altitude_mean_meters | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 952.000000 | 951.000000 | 952.000000 | 830.000000 | 830.000000 | 830.000000 |
| mean | 653.811975 | 192.073529 | 7.557468 | 7.513330 | 7.379338 | 7.533172 | 7.505662 | 7.513214 | 9.839296 | 9.825557 | 9.912384 | 7.483057 | 82.062626 | 0.091292 | 0.380252 | 0.214511 | 4.011555 | 1918.387596 | 1966.069061 | 1942.228329 |
| std | 378.427772 | 120.682457 | 0.400004 | 0.418425 | 0.430553 | 0.403558 | 0.383316 | 0.434140 | 0.584349 | 0.834365 | 0.548040 | 0.469682 | 3.839237 | 0.045773 | 1.658661 | 0.907420 | 5.493412 | 10009.962014 | 10009.287083 | 10009.183888 |
| min | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 |
| 25% | 323.750000 | 50.000000 | 7.420000 | 7.330000 | 7.170000 | 7.330000 | 7.330000 | 7.330000 | 10.000000 | 10.000000 | 10.000000 | 7.250000 | 81.170000 | 0.100000 | 0.000000 | 0.000000 | 1.000000 | 1150.000000 | 1200.000000 | 1200.000000 |
| 50% | 659.500000 | 250.000000 | 7.580000 | 7.580000 | 7.420000 | 7.500000 | 7.500000 | 7.500000 | 10.000000 | 10.000000 | 10.000000 | 7.500000 | 82.500000 | 0.110000 | 0.000000 | 0.000000 | 2.000000 | 1310.640000 | 1350.000000 | 1344.000000 |
| 75% | 972.500000 | 275.000000 | 7.750000 | 7.750000 | 7.580000 | 7.750000 | 7.670000 | 7.750000 | 10.000000 | 10.000000 | 10.000000 | 7.750000 | 83.670000 | 0.120000 | 0.000000 | 0.000000 | 5.000000 | 1600.000000 | 1650.000000 | 1600.000000 |
| max | 1312.000000 | 1062.000000 | 8.750000 | 8.830000 | 8.670000 | 8.750000 | 8.580000 | 8.750000 | 10.000000 | 10.000000 | 10.000000 | 9.250000 | 90.580000 | 0.220000 | 31.000000 | 11.000000 | 55.000000 | 190164.000000 | 190164.000000 | 190164.000000 |
top_coffee_df.columns
Index(['Unnamed: 0', 'Species', 'Owner', 'Country.of.Origin', 'Farm.Name',
'Lot.Number', 'Mill', 'ICO.Number', 'Company', 'Altitude', 'Region',
'Producer', 'Number.of.Bags', 'Bag.Weight', 'In.Country.Partner',
'Harvest.Year', 'Grading.Date', 'Owner.1', 'Variety',
'Processing.Method', 'Aroma', 'Flavor', 'Aftertaste', 'Acidity', 'Body',
'Balance', 'Uniformity', 'Clean.Cup', 'Sweetness', 'Cupper.Points',
'Total.Cup.Points', 'Moisture', 'Category.One.Defects', 'Quakers',
'Color', 'Category.Two.Defects', 'Expiration', 'Certification.Body',
'Certification.Address', 'Certification.Contact', 'unit_of_measurement',
'altitude_low_meters', 'altitude_high_meters', 'altitude_mean_meters'],
dtype='object')
ratings_of_interest = ['Aroma', 'Flavor', 'Aftertaste', 'Acidity', 'Body',
'Balance', ]
coffe_scores_df = top_coffee_df.melt(id_vars='Country.of.Origin',value_vars=ratings_of_interest,
var_name='rating',value_name='score')
coffe_scores_df.head(1)
| Country.of.Origin | rating | score | |
|---|---|---|---|
| 0 | Ethiopia | Aroma | 8.67 |
top_coffee_df.melt(id_vars='Country.of.Origin')['variable'].unique()
array(['Unnamed: 0', 'Species', 'Owner', 'Farm.Name', 'Lot.Number',
'Mill', 'ICO.Number', 'Company', 'Altitude', 'Region', 'Producer',
'Number.of.Bags', 'Bag.Weight', 'In.Country.Partner',
'Harvest.Year', 'Grading.Date', 'Owner.1', 'Variety',
'Processing.Method', 'Aroma', 'Flavor', 'Aftertaste', 'Acidity',
'Body', 'Balance', 'Uniformity', 'Clean.Cup', 'Sweetness',
'Cupper.Points', 'Total.Cup.Points', 'Moisture',
'Category.One.Defects', 'Quakers', 'Color', 'Category.Two.Defects',
'Expiration', 'Certification.Body', 'Certification.Address',
'Certification.Contact', 'unit_of_measurement',
'altitude_low_meters', 'altitude_high_meters',
'altitude_mean_meters'], dtype=object)
top_coffee_df.melt(id_vars='Country.of.Origin',value_vars=ratings_of_interest,)['variable'].unique()
array(['Aroma', 'Flavor', 'Aftertaste', 'Acidity', 'Body', 'Balance'],
dtype=object)
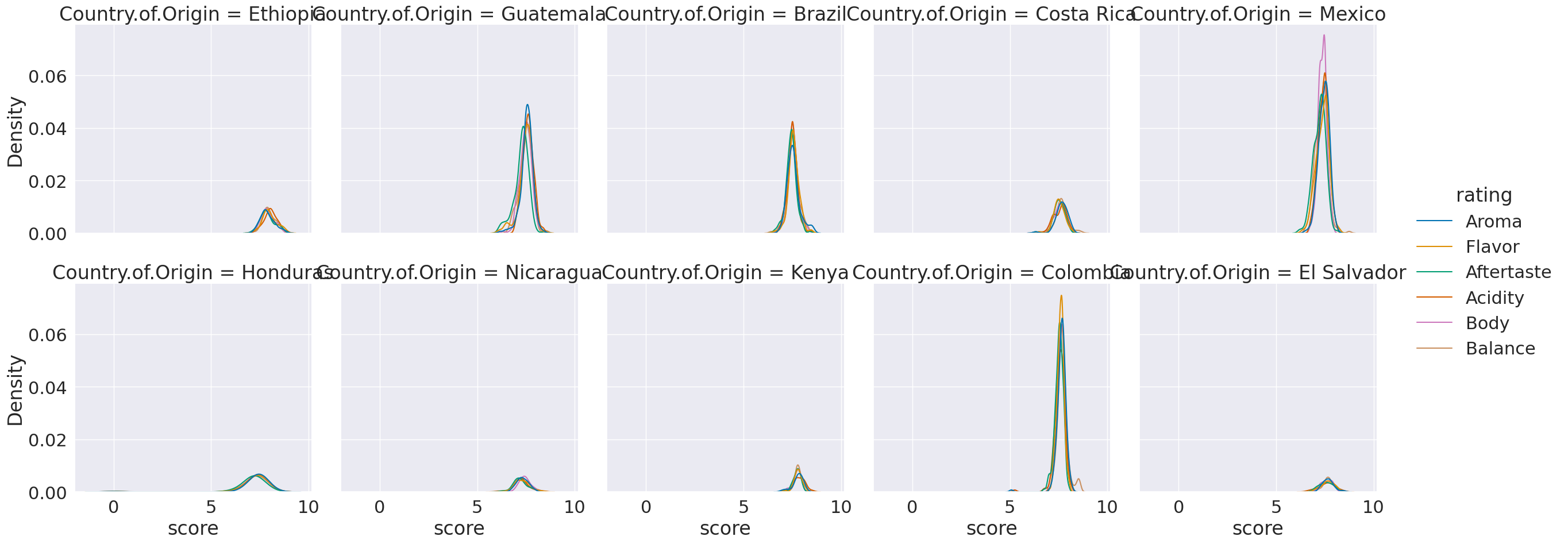
%matplotlib inline
sns.displot(data=coffe_scores_df, x='score',col='Country.of.Origin',
hue = 'rating',col_wrap=5,kind='kde')
<seaborn.axisgrid.FacetGrid at 0x7f78eeaa40d0>
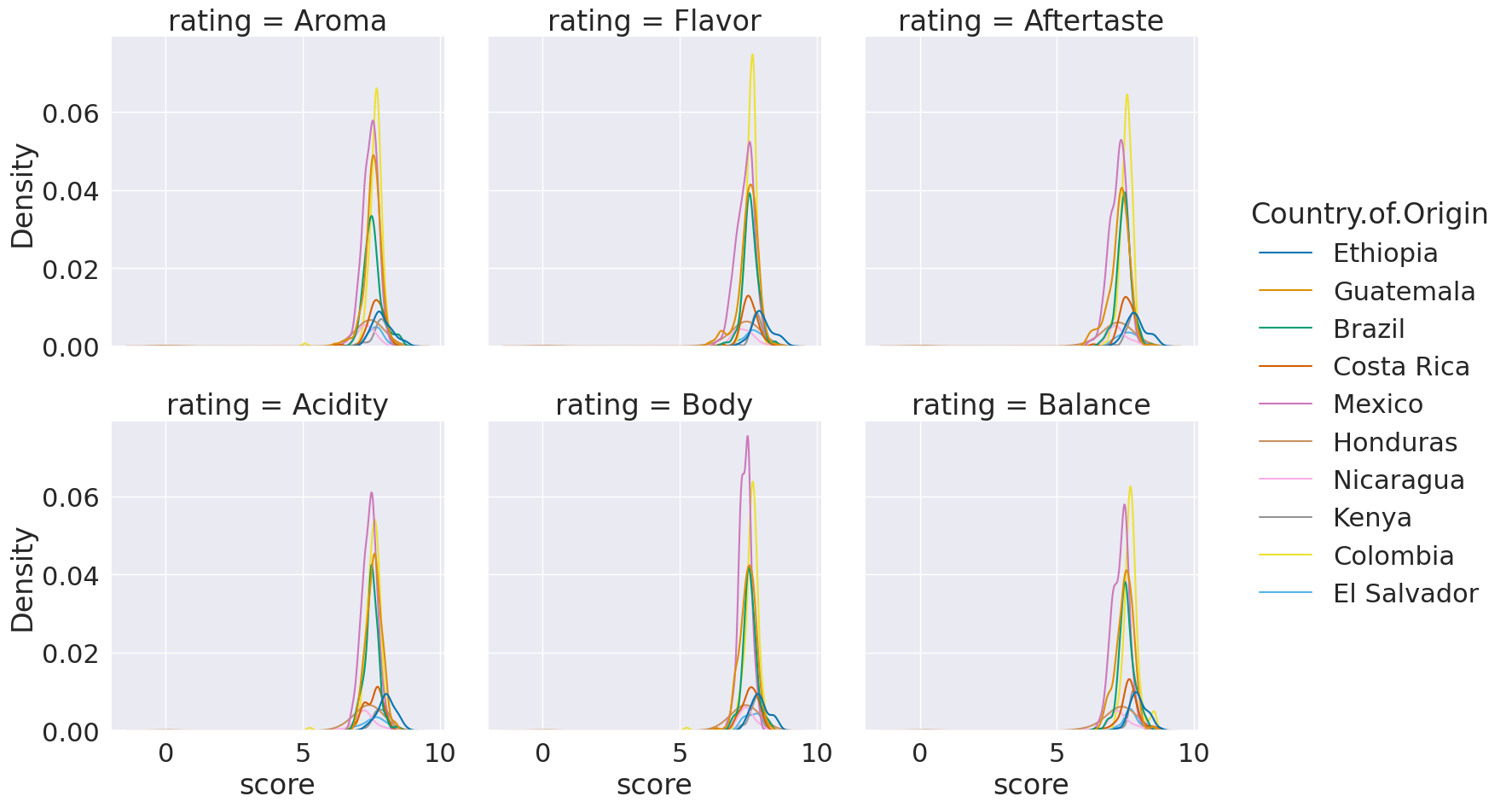
sns.displot(data=coffe_scores_df, x='score',hue='Country.of.Origin',
col = 'rating',col_wrap=3,kind='kde')
<seaborn.axisgrid.FacetGrid at 0x7f7928960d30>
top_coffee_df.columns
Index(['Unnamed: 0', 'Species', 'Owner', 'Country.of.Origin', 'Farm.Name',
'Lot.Number', 'Mill', 'ICO.Number', 'Company', 'Altitude', 'Region',
'Producer', 'Number.of.Bags', 'Bag.Weight', 'In.Country.Partner',
'Harvest.Year', 'Grading.Date', 'Owner.1', 'Variety',
'Processing.Method', 'Aroma', 'Flavor', 'Aftertaste', 'Acidity', 'Body',
'Balance', 'Uniformity', 'Clean.Cup', 'Sweetness', 'Cupper.Points',
'Total.Cup.Points', 'Moisture', 'Category.One.Defects', 'Quakers',
'Color', 'Category.Two.Defects', 'Expiration', 'Certification.Body',
'Certification.Address', 'Certification.Contact', 'unit_of_measurement',
'altitude_low_meters', 'altitude_high_meters', 'altitude_mean_meters'],
dtype='object')
coffe_scores_df2= top_coffee_df.melt(id_vars=['Country.of.Origin','Color'],value_vars=ratings_of_interest,
var_name='rating',value_name='score')
coffe_scores_df2.head(1)
| Country.of.Origin | Color | rating | score | |
|---|---|---|---|---|
| 0 | Ethiopia | Green | Aroma | 8.67 |
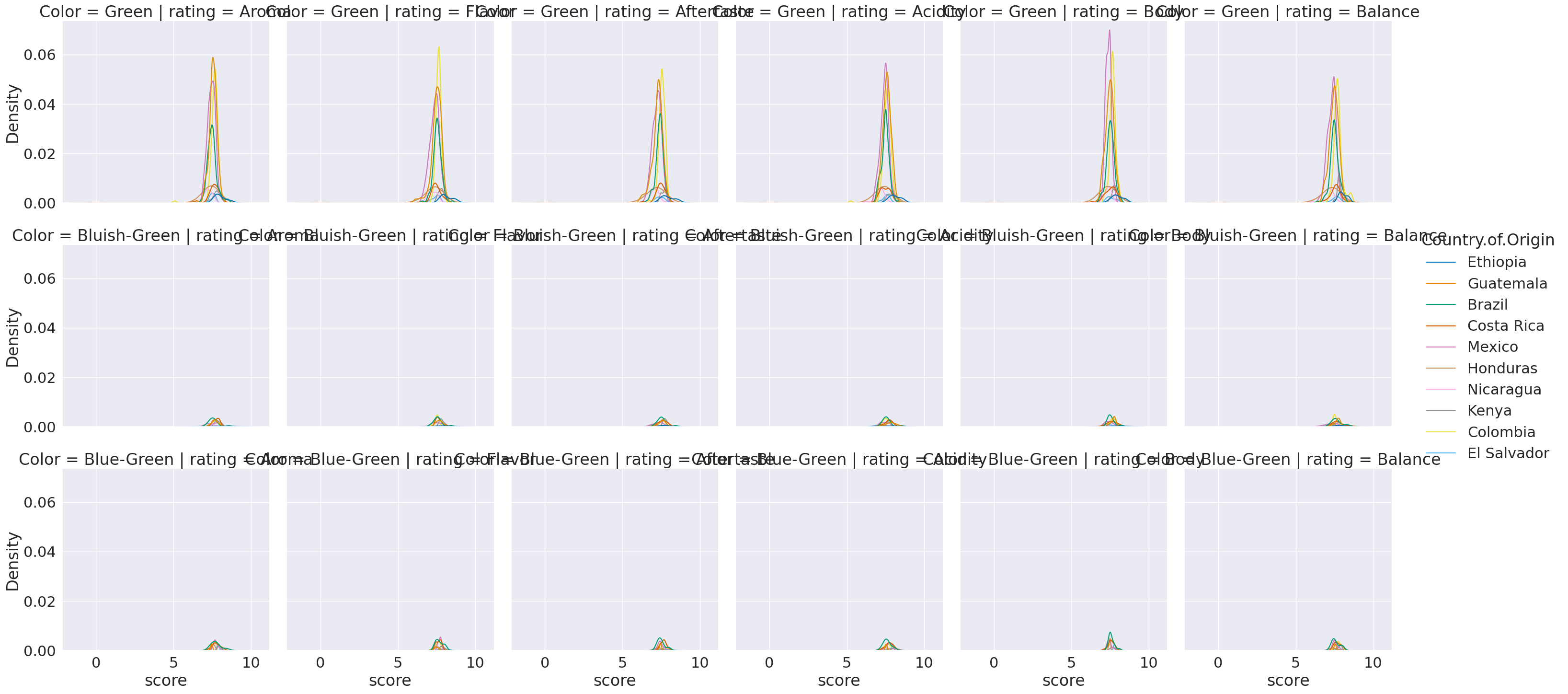
sns.displot(data=coffe_scores_df2, x='score',hue='Country.of.Origin',
col = 'rating',row='Color',kind='kde')
/tmp/ipykernel_1960/3482930274.py:1: UserWarning: Dataset has 0 variance; skipping density estimate. Pass `warn_singular=False` to disable this warning.
sns.displot(data=coffe_scores_df2, x='score',hue='Country.of.Origin',
<seaborn.axisgrid.FacetGrid at 0x7f78edd448b0>
coffee_df.describe()
| Unnamed: 0 | Number.of.Bags | Aroma | Flavor | Aftertaste | Acidity | Body | Balance | Uniformity | Clean.Cup | Sweetness | Cupper.Points | Total.Cup.Points | Moisture | Category.One.Defects | Quakers | Category.Two.Defects | altitude_low_meters | altitude_high_meters | altitude_mean_meters | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.00000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1311.000000 | 1310.000000 | 1311.000000 | 1084.000000 | 1084.000000 | 1084.000000 |
| mean | 656.000763 | 153.887872 | 7.563806 | 7.518070 | 7.397696 | 7.533112 | 7.517727 | 7.517506 | 9.833394 | 9.83312 | 9.903272 | 7.497864 | 82.115927 | 0.088863 | 0.426392 | 0.177099 | 3.591915 | 1759.548954 | 1808.843803 | 1784.196379 |
| std | 378.598733 | 129.733734 | 0.378666 | 0.399979 | 0.405119 | 0.381599 | 0.359213 | 0.406316 | 0.559343 | 0.77135 | 0.530832 | 0.474610 | 3.515761 | 0.047957 | 1.832415 | 0.840583 | 5.350371 | 8767.847252 | 8767.187498 | 8767.016913 |
| min | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.00000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.000000 | 1.000000 | 1.000000 |
| 25% | 328.500000 | 14.500000 | 7.420000 | 7.330000 | 7.250000 | 7.330000 | 7.330000 | 7.330000 | 10.000000 | 10.00000 | 10.000000 | 7.250000 | 81.170000 | 0.090000 | 0.000000 | 0.000000 | 0.000000 | 1100.000000 | 1100.000000 | 1100.000000 |
| 50% | 656.000000 | 175.000000 | 7.580000 | 7.580000 | 7.420000 | 7.500000 | 7.500000 | 7.500000 | 10.000000 | 10.00000 | 10.000000 | 7.500000 | 82.500000 | 0.110000 | 0.000000 | 0.000000 | 2.000000 | 1310.640000 | 1350.000000 | 1310.640000 |
| 75% | 983.500000 | 275.000000 | 7.750000 | 7.750000 | 7.580000 | 7.750000 | 7.670000 | 7.750000 | 10.000000 | 10.00000 | 10.000000 | 7.750000 | 83.670000 | 0.120000 | 0.000000 | 0.000000 | 4.000000 | 1600.000000 | 1650.000000 | 1600.000000 |
| max | 1312.000000 | 1062.000000 | 8.750000 | 8.830000 | 8.670000 | 8.750000 | 8.580000 | 8.750000 | 10.000000 | 10.00000 | 10.000000 | 10.000000 | 90.580000 | 0.280000 | 31.000000 | 11.000000 | 55.000000 | 190164.000000 | 190164.000000 | 190164.000000 |
7.1. More manipulations#
Here, we will make a tiny DataFrame from scratch to illustrate a couple of points
large_num_df = pd.DataFrame(data= [[730000000,392000000,580200000],
[315040009,580000000,967290000]],
columns = ['a','b','c'])
large_num_df
| a | b | c | |
|---|---|---|---|
| 0 | 730000000 | 392000000 | 580200000 |
| 1 | 315040009 | 580000000 | 967290000 |
This dataet is not tidy, but making it this way was faster to set it up. We could make it tidy using melt as is.
large_num_df.melt()
| variable | value | |
|---|---|---|
| 0 | a | 730000000 |
| 1 | a | 315040009 |
| 2 | b | 392000000 |
| 3 | b | 580000000 |
| 4 | c | 580200000 |
| 5 | c | 967290000 |
However, I want an additional variable, so I wil reset the index, which adds an index column for the original index and adds a new index that is numerical. In this case they’re the same.
large_num_df.reset_index()
| index | a | b | c | |
|---|---|---|---|---|
| 0 | 0 | 730000000 | 392000000 | 580200000 |
| 1 | 1 | 315040009 | 580000000 | 967290000 |
If I melt this one, using the index as the id, then I get a reasonable tidy DataFrame
ls_tall_df = large_num_df.reset_index().melt(id_vars='index')
ls_tall_df
| index | variable | value | |
|---|---|---|---|
| 0 | 0 | a | 730000000 |
| 1 | 1 | a | 315040009 |
| 2 | 0 | b | 392000000 |
| 3 | 1 | b | 580000000 |
| 4 | 0 | c | 580200000 |
| 5 | 1 | c | 967290000 |
Now, we can plot.
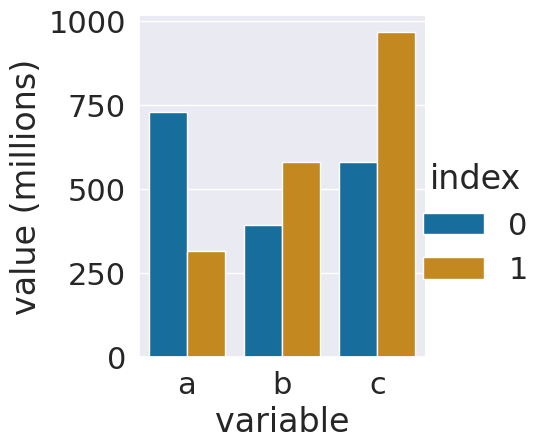
sns.catplot(data = ls_tall_df,x='variable',y='value',
hue='index',kind='bar')
<seaborn.axisgrid.FacetGrid at 0x7f78e8438be0>
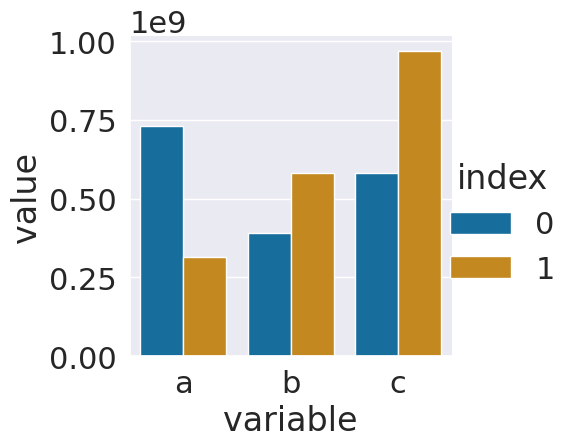
Since the numbers are so big, this might be hard to interpret. Displaying it with all the 0s would not be easier to read. The best thing to do is to add a new colum with adjusted values and a corresponding title.
ls_tall_df['value (millions)'] = ls_tall_df['value']/1000000
ls_tall_df.head()
| index | variable | value | value (millions) | |
|---|---|---|---|---|
| 0 | 0 | a | 730000000 | 730.000000 |
| 1 | 1 | a | 315040009 | 315.040009 |
| 2 | 0 | b | 392000000 | 392.000000 |
| 3 | 1 | b | 580000000 | 580.000000 |
| 4 | 0 | c | 580200000 | 580.200000 |
Now we can plot again, with the smaller values and an updated axis label. Adding a column with the adjusted title is good practice because it does not lose any data and since we set the value and the title at the same time it keeps it clear what the values are.
sns.catplot(data = ls_tall_df,x='variable',y='value (millions)',
hue='index',kind='bar')
<seaborn.axisgrid.FacetGrid at 0x7f78e8e8c970>