Class 6: Exploratory Data Analysis¶
Warmup¶
topics = ['what is data science', 'jupyter', 'conditional','functions', 'lists', 'dictionaries','pandas' ]
topics
['what is data science',
'jupyter',
'conditional',
'functions',
'lists',
'dictionaries',
'pandas']
What happens when we index with -1?
topics[-1]
'pandas'
We get the last value.
Recall last class we used : to index DataFrames with .loc, we can do that with lists too:
topics[-2:]
['dictionaries', 'pandas']
Announcements¶
next assignment will go up today
try practingin throughout the week
Check Brightspace for TA office hours
Why Exploratory Data Analysis (EDA) before cleaning?¶
Typically a Data process looks like one of these figures:

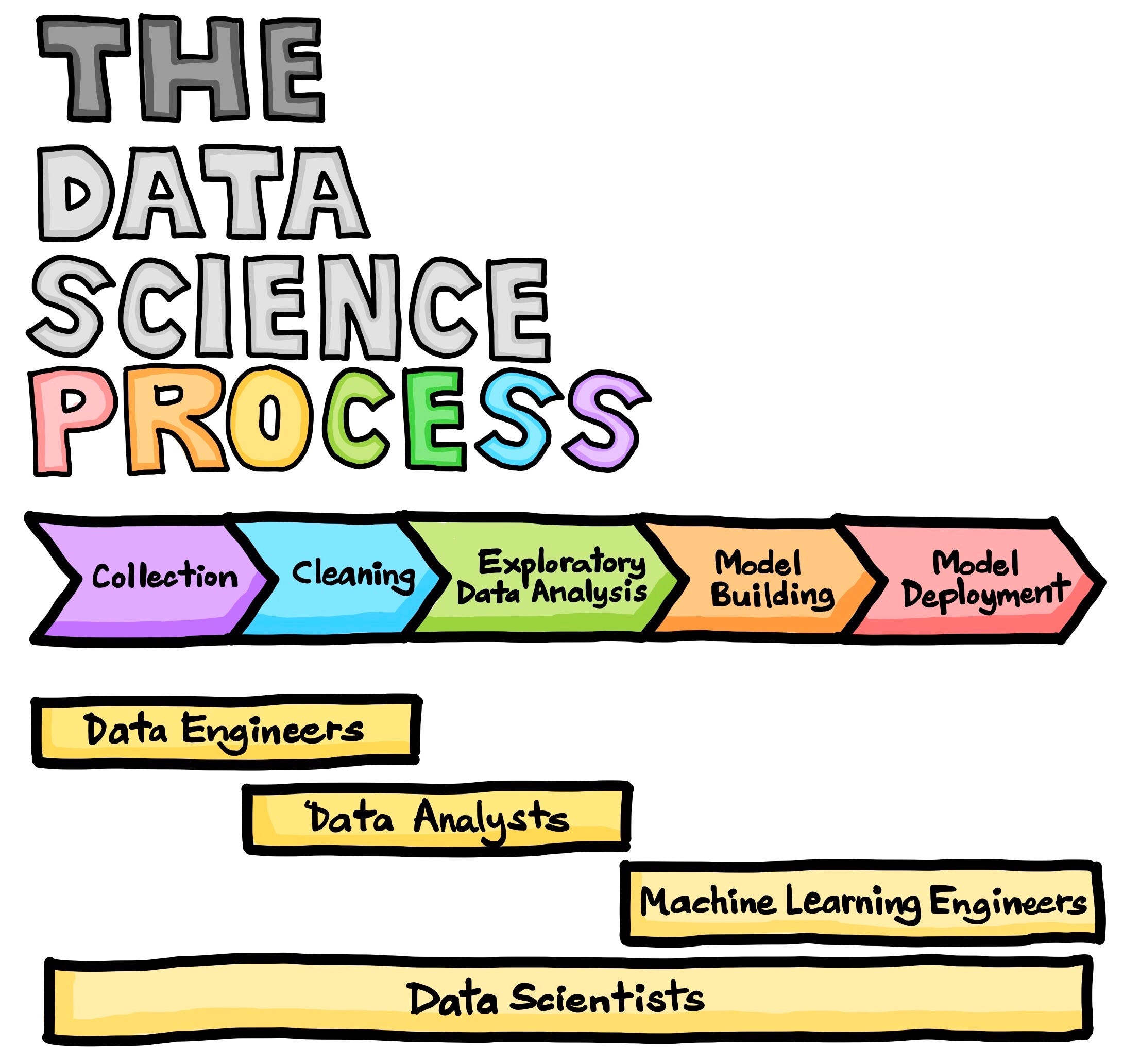
Data cleaning, which some of you asked about on Friday, would typically, happen before EDA, but it’s hard to know what good data looks like, until you’re used to manipulating it. Also, it’s hard to check if data is good (and your cleaning worked) without some EDA. So, I’m choosing to teach EDA first. We’ll do data cleaning next, with these tools from EDA available as options for checking and determining what cleaning to do.
EDA¶
# a version of the SAFI data with more columns
data_url = 'https://raw.githubusercontent.com/brownsarahm/python-socialsci-files/master/data/SAFI_full_shortname.csv'
As usual, we import pandas
import pandas as pd
and pull in the data.
safi_df = pd.read_csv(data_url)
safi_df.head(2)
| key_id | interview_date | quest_no | start | end | province | district | ward | village | years_farm | ... | items_owned | items_owned_other | no_meals | months_lack_food | no_food_mitigation | gps_Latitude | gps_Longitude | gps_Altitude | gps_Accuracy | instanceID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 17 November 2016 | 1 | 2017-03-23T09:49:57.000Z | 2017-04-02T17:29:08.000Z | Manica | Manica | Bandula | God | 11 | ... | ['bicycle' ; 'television' ; 'solar_panel' ; ... | NaN | 2 | ['Jan'] | ['na' ; 'rely_less_food' ; 'reduce_meals' ; ... | -19.112259 | 33.483456 | 698 | 14.0 | uuid:ec241f2c-0609-46ed-b5e8-fe575f6cefef |
| 1 | 2 | 17 November 2016 | 1 | 2017-04-02T09:48:16.000Z | 2017-04-02T17:26:19.000Z | Manica | Manica | Bandula | God | 2 | ... | ['cow_cart' ; 'bicycle' ; 'radio' ; 'cow_pl... | NaN | 2 | ['Jan' ; 'Sept' ; 'Oct' ; 'Nov' ; 'Dec'] | ['na' ; 'reduce_meals' ; 'restrict_adults' ;... | -19.112477 | 33.483416 | 690 | 19.0 | uuid:099de9c9-3e5e-427b-8452-26250e840d6e |
2 rows × 65 columns
Recall on Friday, we can use describe to see several common descriptive statistics
safi_df.describe()
| key_id | quest_no | years_farm | note2 | no_membrs | members_count | years_liv | respondent_wall_type_other | buildings_in_compound | rooms | ... | note | money_source_other | liv_owned_other | liv_count | items_owned_other | no_meals | gps_Latitude | gps_Longitude | gps_Altitude | gps_Accuracy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 131.000000 | 131.000000 | 131.000000 | 0.0 | 131.00000 | 131.00000 | 131.000000 | 0.0 | 131.000000 | 131.000000 | ... | 0.0 | 0.0 | 0.0 | 131.000000 | 0.0 | 131.000000 | 131.000000 | 131.000000 | 131.000000 | 131.000000 |
| mean | 66.000000 | 85.473282 | 15.832061 | NaN | 7.19084 | 7.19084 | 23.053435 | NaN | 2.068702 | 1.740458 | ... | NaN | NaN | NaN | 2.366412 | NaN | 2.603053 | -19.102671 | 33.471971 | 648.221374 | 71.115344 |
| std | 37.960506 | 63.151628 | 10.903883 | NaN | 3.17227 | 3.17227 | 16.913041 | NaN | 1.241530 | 1.092547 | ... | NaN | NaN | NaN | 1.082775 | NaN | 0.491143 | 0.023754 | 0.027081 | 187.697067 | 335.854190 |
| min | 1.000000 | 1.000000 | 1.000000 | NaN | 2.00000 | 2.00000 | 1.000000 | NaN | 1.000000 | 1.000000 | ... | NaN | NaN | NaN | 1.000000 | NaN | 2.000000 | -19.114989 | 33.403836 | 0.000000 | 3.000000 |
| 25% | 33.500000 | 32.500000 | 8.000000 | NaN | 5.00000 | 5.00000 | 12.000000 | NaN | 1.000000 | 1.000000 | ... | NaN | NaN | NaN | 1.000000 | NaN | 2.000000 | -19.112222 | 33.483329 | 691.000000 | 9.000000 |
| 50% | 66.000000 | 66.000000 | 15.000000 | NaN | 7.00000 | 7.00000 | 20.000000 | NaN | 2.000000 | 1.000000 | ... | NaN | NaN | NaN | 2.000000 | NaN | 3.000000 | -19.112188 | 33.483397 | 702.000000 | 11.000000 |
| 75% | 98.500000 | 138.000000 | 20.500000 | NaN | 9.00000 | 9.00000 | 27.500000 | NaN | 3.000000 | 2.000000 | ... | NaN | NaN | NaN | 3.000000 | NaN | 3.000000 | -19.112077 | 33.483438 | 710.000000 | 13.000000 |
| max | 131.000000 | 202.000000 | 60.000000 | NaN | 19.00000 | 19.00000 | 96.000000 | NaN | 8.000000 | 8.000000 | ... | NaN | NaN | NaN | 5.000000 | NaN | 3.000000 | -19.042909 | 33.488268 | 745.000000 | 2099.999000 |
8 rows × 25 columns
Then we remember that this includes key_id as a variable tht we don’t actually want to treat as a variable, it’s an index. We can change that with set_index and we can do it in memory (without assignment) using the inplace keyword.
safi_df.set_index('key_id',inplace=True)
safi_df.describe()
| quest_no | years_farm | note2 | no_membrs | members_count | years_liv | respondent_wall_type_other | buildings_in_compound | rooms | no_plots | ... | note | money_source_other | liv_owned_other | liv_count | items_owned_other | no_meals | gps_Latitude | gps_Longitude | gps_Altitude | gps_Accuracy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 131.000000 | 131.000000 | 0.0 | 131.00000 | 131.00000 | 131.000000 | 0.0 | 131.000000 | 131.000000 | 131.000000 | ... | 0.0 | 0.0 | 0.0 | 131.000000 | 0.0 | 131.000000 | 131.000000 | 131.000000 | 131.000000 | 131.000000 |
| mean | 85.473282 | 15.832061 | NaN | 7.19084 | 7.19084 | 23.053435 | NaN | 2.068702 | 1.740458 | 2.229008 | ... | NaN | NaN | NaN | 2.366412 | NaN | 2.603053 | -19.102671 | 33.471971 | 648.221374 | 71.115344 |
| std | 63.151628 | 10.903883 | NaN | 3.17227 | 3.17227 | 16.913041 | NaN | 1.241530 | 1.092547 | 1.078210 | ... | NaN | NaN | NaN | 1.082775 | NaN | 0.491143 | 0.023754 | 0.027081 | 187.697067 | 335.854190 |
| min | 1.000000 | 1.000000 | NaN | 2.00000 | 2.00000 | 1.000000 | NaN | 1.000000 | 1.000000 | 1.000000 | ... | NaN | NaN | NaN | 1.000000 | NaN | 2.000000 | -19.114989 | 33.403836 | 0.000000 | 3.000000 |
| 25% | 32.500000 | 8.000000 | NaN | 5.00000 | 5.00000 | 12.000000 | NaN | 1.000000 | 1.000000 | 2.000000 | ... | NaN | NaN | NaN | 1.000000 | NaN | 2.000000 | -19.112222 | 33.483329 | 691.000000 | 9.000000 |
| 50% | 66.000000 | 15.000000 | NaN | 7.00000 | 7.00000 | 20.000000 | NaN | 2.000000 | 1.000000 | 2.000000 | ... | NaN | NaN | NaN | 2.000000 | NaN | 3.000000 | -19.112188 | 33.483397 | 702.000000 | 11.000000 |
| 75% | 138.000000 | 20.500000 | NaN | 9.00000 | 9.00000 | 27.500000 | NaN | 3.000000 | 2.000000 | 3.000000 | ... | NaN | NaN | NaN | 3.000000 | NaN | 3.000000 | -19.112077 | 33.483438 | 710.000000 | 13.000000 |
| max | 202.000000 | 60.000000 | NaN | 19.00000 | 19.00000 | 96.000000 | NaN | 8.000000 | 8.000000 | 8.000000 | ... | NaN | NaN | NaN | 5.000000 | NaN | 3.000000 | -19.042909 | 33.488268 | 745.000000 | 2099.999000 |
8 rows × 24 columns
Try it yourself!
You can use settings on pd.read_csv to set the index when the data is read in instead of doing this after the fact
We can also use the descriptive statistics on a single variable.
safi_df['years_farm'].describe()
count 131.000000
mean 15.832061
std 10.903883
min 1.000000
25% 8.000000
50% 15.000000
75% 20.500000
max 60.000000
Name: years_farm, dtype: float64
Note however that this is not well formatted, that’s because it’s a Series instead of a DataFrame
type(safi_df['years_farm'].describe())
pandas.core.series.Series
We can use reset_index() to make it back into a dataframe
safi_df['years_farm'].describe().reset_index()
| index | years_farm | |
|---|---|---|
| 0 | count | 131.000000 |
| 1 | mean | 15.832061 |
| 2 | std | 10.903883 |
| 3 | min | 1.000000 |
| 4 | 25% | 8.000000 |
| 5 | 50% | 15.000000 |
| 6 | 75% | 20.500000 |
| 7 | max | 60.000000 |
And we can drop the added index:
safi_df['years_farm'].describe().reset_index().set_index('index')
| years_farm | |
|---|---|
| index | |
| count | 131.000000 |
| mean | 15.832061 |
| std | 10.903883 |
| min | 1.000000 |
| 25% | 8.000000 |
| 50% | 15.000000 |
| 75% | 20.500000 |
| max | 60.000000 |
Note
See that we can chain operations together. This is a helpful feature, but to be used with care. Pep8, the style conventions for python, recommends no more than 79 characters per line. A summary and another summary
We can also use individual summary statistics on DataFrame or Series objects
safi_df['no_membrs'].max()
19
Split - Apply - Combine¶
A powerful tool in pandas (and data analysis in general) is the ability to apply functions to subsets of the data.
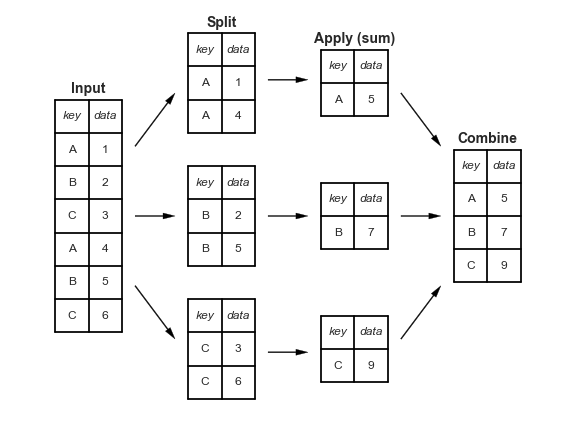
For example, we can get descriptive statistics per village
safi_df.groupby('village').describe()
| quest_no | years_farm | ... | gps_Altitude | gps_Accuracy | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | count | mean | ... | 75% | max | count | mean | std | min | 25% | 50% | 75% | max | |
| village | |||||||||||||||||||||
| Chirodzo | 39.0 | 62.487179 | 44.261705 | 8.0 | 44.5 | 55.0 | 64.5 | 200.0 | 39.0 | 17.282051 | ... | 710.0 | 733.0 | 39.0 | 12.461538 | 7.089080 | 4.0 | 9.0 | 11.0 | 12.0 | 39.000 |
| God | 43.0 | 81.720930 | 77.863839 | 1.0 | 14.5 | 40.0 | 168.5 | 202.0 | 43.0 | 14.883721 | ... | 709.5 | 722.0 | 43.0 | 12.630512 | 6.075239 | 4.0 | 9.0 | 11.0 | 13.5 | 30.000 |
| Ruaca | 49.0 | 107.061224 | 55.024013 | 23.0 | 72.0 | 113.0 | 152.0 | 194.0 | 49.0 | 15.510204 | ... | 711.0 | 745.0 | 49.0 | 169.122408 | 538.291474 | 3.0 | 6.0 | 9.0 | 12.0 | 2099.999 |
3 rows × 192 columns
We can also rearrange this into a more usable format than this very wide format:
safi_df.groupby('village').describe().unstack().reset_index()
| level_0 | level_1 | village | 0 | |
|---|---|---|---|---|
| 0 | quest_no | count | Chirodzo | 39.000000 |
| 1 | quest_no | count | God | 43.000000 |
| 2 | quest_no | count | Ruaca | 49.000000 |
| 3 | quest_no | mean | Chirodzo | 62.487179 |
| 4 | quest_no | mean | God | 81.720930 |
| ... | ... | ... | ... | ... |
| 571 | gps_Accuracy | 75% | God | 13.500000 |
| 572 | gps_Accuracy | 75% | Ruaca | 12.000000 |
| 573 | gps_Accuracy | max | Chirodzo | 39.000000 |
| 574 | gps_Accuracy | max | God | 30.000000 |
| 575 | gps_Accuracy | max | Ruaca | 2099.999000 |
576 rows × 4 columns
This, however, gives some funny variable names, to fix that, we first save it to a variable.
village_summary_df = safi_df.groupby('village').describe().unstack().reset_index()
Now we can use the rename function
help(village_summary_df.rename)
Help on method rename in module pandas.core.frame:
rename(mapper=None, index=None, columns=None, axis=None, copy=True, inplace=False, level=None, errors='ignore') method of pandas.core.frame.DataFrame instance
Alter axes labels.
Function / dict values must be unique (1-to-1). Labels not contained in
a dict / Series will be left as-is. Extra labels listed don't throw an
error.
See the :ref:`user guide <basics.rename>` for more.
Parameters
----------
mapper : dict-like or function
Dict-like or function transformations to apply to
that axis' values. Use either ``mapper`` and ``axis`` to
specify the axis to target with ``mapper``, or ``index`` and
``columns``.
index : dict-like or function
Alternative to specifying axis (``mapper, axis=0``
is equivalent to ``index=mapper``).
columns : dict-like or function
Alternative to specifying axis (``mapper, axis=1``
is equivalent to ``columns=mapper``).
axis : {0 or 'index', 1 or 'columns'}, default 0
Axis to target with ``mapper``. Can be either the axis name
('index', 'columns') or number (0, 1). The default is 'index'.
copy : bool, default True
Also copy underlying data.
inplace : bool, default False
Whether to return a new DataFrame. If True then value of copy is
ignored.
level : int or level name, default None
In case of a MultiIndex, only rename labels in the specified
level.
errors : {'ignore', 'raise'}, default 'ignore'
If 'raise', raise a `KeyError` when a dict-like `mapper`, `index`,
or `columns` contains labels that are not present in the Index
being transformed.
If 'ignore', existing keys will be renamed and extra keys will be
ignored.
Returns
-------
DataFrame or None
DataFrame with the renamed axis labels or None if ``inplace=True``.
Raises
------
KeyError
If any of the labels is not found in the selected axis and
"errors='raise'".
See Also
--------
DataFrame.rename_axis : Set the name of the axis.
Examples
--------
``DataFrame.rename`` supports two calling conventions
* ``(index=index_mapper, columns=columns_mapper, ...)``
* ``(mapper, axis={'index', 'columns'}, ...)``
We *highly* recommend using keyword arguments to clarify your
intent.
Rename columns using a mapping:
>>> df = pd.DataFrame({"A": [1, 2, 3], "B": [4, 5, 6]})
>>> df.rename(columns={"A": "a", "B": "c"})
a c
0 1 4
1 2 5
2 3 6
Rename index using a mapping:
>>> df.rename(index={0: "x", 1: "y", 2: "z"})
A B
x 1 4
y 2 5
z 3 6
Cast index labels to a different type:
>>> df.index
RangeIndex(start=0, stop=3, step=1)
>>> df.rename(index=str).index
Index(['0', '1', '2'], dtype='object')
>>> df.rename(columns={"A": "a", "B": "b", "C": "c"}, errors="raise")
Traceback (most recent call last):
KeyError: ['C'] not found in axis
Using axis-style parameters:
>>> df.rename(str.lower, axis='columns')
a b
0 1 4
1 2 5
2 3 6
>>> df.rename({1: 2, 2: 4}, axis='index')
A B
0 1 4
2 2 5
4 3 6
Rename takes a dict-like or function that maps from what’s there to what to change it to. We can either provide the mapper and an axis or pass the mapper as the columns parameter. The columns parameter makes it more human readable, and explicit what we’re doing.
renaming_dict = {'level_0':'variable',
'level_1':'statistic',
0:'value'}
village_summary_df.rename(columns= renaming_dict,inplace=True)
Now we have a much better summary table. This is in what’s called tidy format. Each colum
village_summary_df.head()
| variable | statistic | village | value | |
|---|---|---|---|---|
| 0 | quest_no | count | Chirodzo | 39.000000 |
| 1 | quest_no | count | God | 43.000000 |
| 2 | quest_no | count | Ruaca | 49.000000 |
| 3 | quest_no | mean | Chirodzo | 62.487179 |
| 4 | quest_no | mean | God | 81.720930 |
How could we use this to compute the average across villages for each statistic of each variable?
Try it yourself!
What would it look like to do this from the first result of describe() on the groupby, instead of with the unstack and reset_index?
We do need to groupby 'statistic' and use mean, but that’s not enough:
village_summary_df.groupby('statistic').mean()
| value | |
|---|---|
| statistic | |
| 25% | 44.807522 |
| 50% | 48.428030 |
| 75% | 54.062308 |
| count | 30.388889 |
| max | 105.468983 |
| mean | 49.868824 |
| min | 13.756913 |
| std | 24.588967 |
This averages across all of the variables, too. So instead we need to groupby two variables.
village_summary_df.groupby(['statistic','variable']).mean()
| value | ||
|---|---|---|
| statistic | variable | |
| 25% | buildings_in_compound | 1.000000 |
| gps_Accuracy | 8.000000 | |
| gps_Altitude | 691.833333 | |
| gps_Latitude | -19.112221 | |
| gps_Longitude | 33.480944 | |
| ... | ... | ... |
| std | respondent_wall_type_other | NaN |
| rooms | 1.082409 | |
| years_farm | 10.832121 | |
| years_liv | 16.549343 | |
| yes_group_count | 1.074600 |
192 rows × 1 columns
safi_df.columns
Index(['interview_date', 'quest_no', 'start', 'end', 'province', 'district',
'ward', 'village', 'years_farm', 'agr_assoc', 'note2', 'no_membrs',
'members_count', 'remittance_money', 'years_liv', 'parents_liv',
'sp_parents_liv', 'grand_liv', 'sp_grand_liv', 'respondent_roof_type',
'respondent_wall_type', 'respondent_wall_type_other',
'respondent_floor_type', 'window_type', 'buildings_in_compound',
'rooms', 'other_buildings', 'no_plots', 'plots_count', 'water_use',
'no_group_count', 'yes_group_count', 'no_enough_water',
'months_no_water', 'period_use', 'exper_other', 'other_meth',
'res_change', 'memb_assoc', 'resp_assoc', 'fees_water',
'affect_conflicts', 'note', 'need_money', 'money_source',
'money_source_other', 'crops_contr', 'emply_lab', 'du_labour',
'liv_owned', 'liv_owned_other', 'liv_count', 'poultry',
'du_look_aftr_cows', 'items_owned', 'items_owned_other', 'no_meals',
'months_lack_food', 'no_food_mitigation', 'gps_Latitude',
'gps_Longitude', 'gps_Altitude', 'gps_Accuracy', 'instanceID'],
dtype='object')
What is the most common combination of respondent_wall_type and respondent_floor_type?
safi_df.groupby(['respondent_wall_type','respondent_floor_type'])['instanceID'].count().reset_index()
| respondent_wall_type | respondent_floor_type | instanceID | |
|---|---|---|---|
| 0 | burntbricks | cement | 30 |
| 1 | burntbricks | earth | 37 |
| 2 | cement | cement | 1 |
| 3 | muddaub | cement | 3 |
| 4 | muddaub | earth | 43 |
| 5 | sunbricks | cement | 4 |
| 6 | sunbricks | earth | 13 |
We can read this table to see the result. Or we might want it in a different way
safi_df.groupby(['respondent_wall_type','respondent_floor_type'])['instanceID'].count().unstack()
| respondent_floor_type | cement | earth |
|---|---|---|
| respondent_wall_type | ||
| burntbricks | 30.0 | 37.0 |
| cement | 1.0 | NaN |
| muddaub | 3.0 | 43.0 |
| sunbricks | 4.0 | 13.0 |
Questions After Class¶
What does the inplace parameter do?¶
We used inplace for the first time today right after we read in the data. Let’s make a new DataFrame and compare what happens with and without.
safi_df_noinplace = pd.read_csv(data_url)
safi_df_inplace = pd.read_csv(data_url)
type(safi_df_noinplace.set_index('key_id'))
pandas.core.frame.DataFrame
safi_df_noinplace.head()
| key_id | interview_date | quest_no | start | end | province | district | ward | village | years_farm | ... | items_owned | items_owned_other | no_meals | months_lack_food | no_food_mitigation | gps_Latitude | gps_Longitude | gps_Altitude | gps_Accuracy | instanceID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 17 November 2016 | 1 | 2017-03-23T09:49:57.000Z | 2017-04-02T17:29:08.000Z | Manica | Manica | Bandula | God | 11 | ... | ['bicycle' ; 'television' ; 'solar_panel' ; ... | NaN | 2 | ['Jan'] | ['na' ; 'rely_less_food' ; 'reduce_meals' ; ... | -19.112259 | 33.483456 | 698 | 14.0 | uuid:ec241f2c-0609-46ed-b5e8-fe575f6cefef |
| 1 | 2 | 17 November 2016 | 1 | 2017-04-02T09:48:16.000Z | 2017-04-02T17:26:19.000Z | Manica | Manica | Bandula | God | 2 | ... | ['cow_cart' ; 'bicycle' ; 'radio' ; 'cow_pl... | NaN | 2 | ['Jan' ; 'Sept' ; 'Oct' ; 'Nov' ; 'Dec'] | ['na' ; 'reduce_meals' ; 'restrict_adults' ;... | -19.112477 | 33.483416 | 690 | 19.0 | uuid:099de9c9-3e5e-427b-8452-26250e840d6e |
| 2 | 3 | 17 November 2016 | 3 | 2017-04-02T14:35:26.000Z | 2017-04-02T17:26:53.000Z | Manica | Manica | Bandula | God | 40 | ... | ['solar_torch'] | NaN | 2 | ['Jan' ; 'Feb' ; 'Mar' ; 'Oct' ; 'Nov' ; ... | ['na' ; 'restrict_adults' ; 'lab_ex_food'] | -19.112108 | 33.483450 | 674 | 13.0 | uuid:193d7daf-9582-409b-bf09-027dd36f9007 |
| 3 | 4 | 17 November 2016 | 4 | 2017-04-02T14:55:18.000Z | 2017-04-02T17:27:16.000Z | Manica | Manica | Bandula | God | 6 | ... | ['bicycle' ; 'radio' ; 'cow_plough' ; 'sola... | NaN | 2 | ['Sept' ; 'Oct' ; 'Nov' ; 'Dec'] | ['na' ; 'reduce_meals' ; 'restrict_adults' ;... | -19.112229 | 33.483424 | 679 | 5.0 | uuid:148d1105-778a-4755-aa71-281eadd4a973 |
| 4 | 5 | 17 November 2016 | 5 | 2017-04-02T15:10:35.000Z | 2017-04-02T17:27:35.000Z | Manica | Manica | Bandula | God | 18 | ... | ['motorcyle' ; 'radio' ; 'cow_plough' ; 'mo... | NaN | 2 | ['Aug' ; 'Sept' ; 'Oct' ; 'Nov'] | ['na' ; 'go_forest' ; 'migrate'] | -19.112217 | 33.483425 | 689 | 10.0 | uuid:2c867811-9696-4966-9866-f35c3e97d02d |
5 rows × 65 columns
type(safi_df_inplace.set_index('key_id',inplace=True))
NoneType
safi_df_inplace.head()
| interview_date | quest_no | start | end | province | district | ward | village | years_farm | agr_assoc | ... | items_owned | items_owned_other | no_meals | months_lack_food | no_food_mitigation | gps_Latitude | gps_Longitude | gps_Altitude | gps_Accuracy | instanceID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| key_id | |||||||||||||||||||||
| 1 | 17 November 2016 | 1 | 2017-03-23T09:49:57.000Z | 2017-04-02T17:29:08.000Z | Manica | Manica | Bandula | God | 11 | no | ... | ['bicycle' ; 'television' ; 'solar_panel' ; ... | NaN | 2 | ['Jan'] | ['na' ; 'rely_less_food' ; 'reduce_meals' ; ... | -19.112259 | 33.483456 | 698 | 14.0 | uuid:ec241f2c-0609-46ed-b5e8-fe575f6cefef |
| 2 | 17 November 2016 | 1 | 2017-04-02T09:48:16.000Z | 2017-04-02T17:26:19.000Z | Manica | Manica | Bandula | God | 2 | yes | ... | ['cow_cart' ; 'bicycle' ; 'radio' ; 'cow_pl... | NaN | 2 | ['Jan' ; 'Sept' ; 'Oct' ; 'Nov' ; 'Dec'] | ['na' ; 'reduce_meals' ; 'restrict_adults' ;... | -19.112477 | 33.483416 | 690 | 19.0 | uuid:099de9c9-3e5e-427b-8452-26250e840d6e |
| 3 | 17 November 2016 | 3 | 2017-04-02T14:35:26.000Z | 2017-04-02T17:26:53.000Z | Manica | Manica | Bandula | God | 40 | no | ... | ['solar_torch'] | NaN | 2 | ['Jan' ; 'Feb' ; 'Mar' ; 'Oct' ; 'Nov' ; ... | ['na' ; 'restrict_adults' ; 'lab_ex_food'] | -19.112108 | 33.483450 | 674 | 13.0 | uuid:193d7daf-9582-409b-bf09-027dd36f9007 |
| 4 | 17 November 2016 | 4 | 2017-04-02T14:55:18.000Z | 2017-04-02T17:27:16.000Z | Manica | Manica | Bandula | God | 6 | no | ... | ['bicycle' ; 'radio' ; 'cow_plough' ; 'sola... | NaN | 2 | ['Sept' ; 'Oct' ; 'Nov' ; 'Dec'] | ['na' ; 'reduce_meals' ; 'restrict_adults' ;... | -19.112229 | 33.483424 | 679 | 5.0 | uuid:148d1105-778a-4755-aa71-281eadd4a973 |
| 5 | 17 November 2016 | 5 | 2017-04-02T15:10:35.000Z | 2017-04-02T17:27:35.000Z | Manica | Manica | Bandula | God | 18 | no | ... | ['motorcyle' ; 'radio' ; 'cow_plough' ; 'mo... | NaN | 2 | ['Aug' ; 'Sept' ; 'Oct' ; 'Nov'] | ['na' ; 'go_forest' ; 'migrate'] | -19.112217 | 33.483425 | 689 | 10.0 | uuid:2c867811-9696-4966-9866-f35c3e97d02d |
5 rows × 64 columns
Without inplace, set_index returns a dataframe, with the index changed, with inplace it returns None. But, without inplace, we don’t see the desired effect when we do our next step, but with inplace the index is changed.
Without the inplace, to save the changes you need to use an assignment. The following is equivalent to using inplace.
safi_df_noinplace= safi_df_noinplace.set_index('key_id'))
safi_df_noinplace.head()
File "<ipython-input-30-1a84c65392b3>", line 1
safi_df_noinplace= safi_df_noinplace.set_index('key_id'))
^
SyntaxError: invalid syntax
Why is a for loop slower than a pandas operation?¶
TL;DR: for this example, it’s actually possible to relatively easily write a faster loop, but for other operations, it’s unlikely.
Basically, the advantage of using the library functions is that someone has already put a lot of though into the optimal way to implement things and it leverages core features of the data structure. Here is a blog post about iterating and applying functions in pandas. It covers topics we haven’t yet seen, but will.
The pandas implementation is also fewer lines and can scale for large datasets eaisly.
To test, we can do an experiment. Let’s take the last thing we covered in class today, finding the most common combination of floor and wall types.
First we’ll look at two solutions and verify that they’re the same.
First, how to do it with pandas
wall_floor_df = safi_df.groupby(['respondent_wall_type','respondent_floor_type'])['instanceID'].count()
wall_floor_df
respondent_wall_type respondent_floor_type
burntbricks cement 30
earth 37
cement cement 1
muddaub cement 3
earth 43
sunbricks cement 4
earth 13
Name: instanceID, dtype: int64
We can read the answer off of that table, but let’s get it programmatically:
wall_floor_df.idxmax()
('muddaub', 'earth')
Next, how to do it with a for loop (if you have a better for loop, make a PR so share it).
wall_floor_dict = {}
for wall,floor in zip(safi_df['respondent_wall_type'],safi_df['respondent_floor_type']):
wf_key = '_'.join([wall,floor])
if wf_key in wall_floor_dict.keys():
wall_floor_dict[wf_key] +=1
else:
wall_floor_dict[wf_key] =1
wall_floor_dict
{'muddaub_earth': 43,
'burntbricks_cement': 30,
'burntbricks_earth': 37,
'sunbricks_earth': 13,
'muddaub_cement': 3,
'sunbricks_cement': 4,
'cement_cement': 1}
Again, we can read from the dict, but lets find it
max(wall_floor_dict, key=wall_floor_dict.get).split('_')
['muddaub', 'earth']
Now we can use a special feature of Jupyter notebooks to time them and see which is faster, called the %timeit magic. Displaying visual things is always slow, so we’ll take the line that would display out of both options.
Now timing the pandas way, with a human deciding
%%timeit -o
wall_floor_df = safi_df.groupby(['respondent_wall_type','respondent_floor_type'])['instanceID'].count().reset_index()
2.31 ms ± 60.8 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
<TimeitResult : 2.31 ms ± 60.8 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)>
t_pandas = _
And capture the time by using the _ it gets the std out from the previous cell.
For loop with required interpretation
%%timeit -o
wall_floor_dict = {}
for wall,floor in zip(safi_df['respondent_wall_type'],safi_df['respondent_floor_type']):
wf_key = '_'.join([wall,floor])
if wf_key in wall_floor_dict.keys():
wall_floor_dict[wf_key] +=1
else:
wall_floor_dict[wf_key] =1
max(wall_floor_dict, key=wall_floor_dict.get).split('_')
87.1 µs ± 801 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)
<TimeitResult : 87.1 µs ± 801 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)>
t_loop = _
On my local computer it was about 12x faster with that loop. Here, we’ll calculate it for the GitHub servers that host the course manual.
t_pandas.average/t_loop.average
26.57731776507862
For a more a complex question (say we wanted to know a statistic more complicated than the count) the loop gets harder (and probably slower), but the pandas operation stays about the same.
If you’ve made it this far, let me know how you found these notes.
