Interpetting and Evaluating Naive Bayes
Contents
16. Interpetting and Evaluating Naive Bayes#
We’ll pikc up where we left off on Wedneday and we’ll cover what the Naive Bayes model assumes and some more evaluation. Next week, we’ll see a bit more about it’s predictions and see a new classifier.
import pandas as pd
import seaborn as sns
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import confusion_matrix, classification_report
iris_df = sns.load_dataset('iris')
sns.set_theme(palette= "colorblind")
Again we’ll load the data and split it to test and train and indicate which variables to use as feature and the target.
feature_vars = ['sepal_length', 'sepal_width','petal_length', 'petal_width',]
target_var = 'species'
X_train, X_test, y_train, y_test = train_test_split(iris_df[feature_vars],
iris_df[target_var],
test_size=.5, random_state=0)
This time we also made the test size larger (50% instead of default 25%)
16.1. What does Naive Bayes do?#
Gaussian = features distributed according to the Gaussian Distribution (normal curve) Naive = indepdent features Bayes = most probable
To see, first we’ll fit the model, hten examine it’s strucutre.
gnb= GaussianNB()
gnb.fit(X_train,y_train)
gnb.__dict__
{'priors': None,
'var_smoothing': 1e-09,
'classes_': array(['setosa', 'versicolor', 'virginica'], dtype='<U10'),
'feature_names_in_': array(['sepal_length', 'sepal_width', 'petal_length', 'petal_width'],
dtype=object),
'n_features_in_': 4,
'epsilon_': 3.6399039999999994e-09,
'theta_': array([[4.97586207, 3.35862069, 1.44827586, 0.23448276],
[5.935 , 2.71 , 4.185 , 1.3 ],
[6.77692308, 3.09230769, 5.73461538, 2.10769231]]),
'var_': array([[0.10321047, 0.13208086, 0.01629013, 0.00846612],
[0.256275 , 0.0829 , 0.255275 , 0.046 ],
[0.38869823, 0.10147929, 0.31303255, 0.04763314]]),
'class_count_': array([29., 20., 26.]),
'class_prior_': array([0.38666667, 0.26666667, 0.34666667])}
The attributes of the estimator object (gbn) describe the data (eg the class list) and the model’s parameters. The theta_ (\(\theta\))
represents the mean and the sigma_ (\(\sigma\)) represents the variance of the
distributions.
Try it Yourself
Could you use what we learned about EDA to find the mean and variance of each feature for each species of flower?
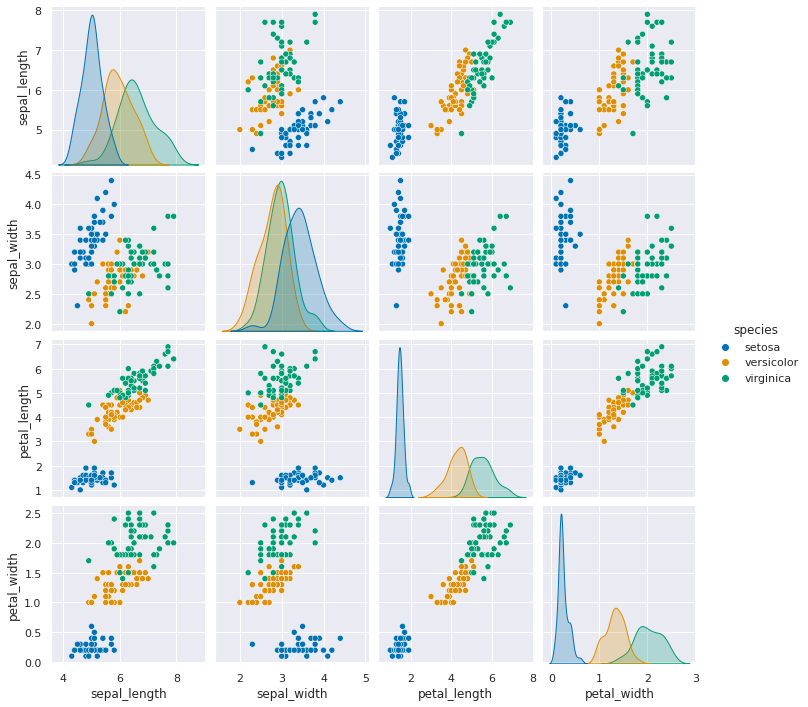
When we look at the data with a pair plot we see the distriution. This data is not perfectly Gaussian, but it’s pretty close to unimodal (one peak) for each feature and they’re relatively normal curve like (as opposed to a vary sharp spike like a laplacian )
sns.pairplot(data=iris_df, hue='species')
<seaborn.axisgrid.PairGrid at 0x7fed5c38c0d0>
Because the GaussianNB classifier calculates parameters that describe the assumed distribuiton of the data is is called a generative classifier. From a generative classifer, we can generate synthetic data that is from the distribution the classifer learned. If this data looks like our real data, then the model assumptions fit well.
Warning
the details of this math are not required understanding, but this describes the following block of code
To do this, we extract the mean and variance parameters from the model
(gnb.theta_,gnb.sigma_) and zip them together to create an iterable object
that in each iteration returns one value from each list (for th, sig in zip(gnb.theta_,gnb.sigma_)).
We do this inside of a list comprehension and for each th,sig where th is
from gnb.theta_ and sig is from gnb.sigma_ we use np.random.multivariate_normal
to get 20 samples. In a general multivariate normal distribution the second parameter is actually a covariance
matrix. This describes both the variance of each individual feature and the
correlation of the features. Since Naive Bayes is Naive it assumes the features
are independent or have 0 correlation. So, to create the matrix from the vector
of variances we multiply by np.eye(4) which is the identity matrix or a matrix
with 1 on the diagonal and 0 elsewhere. Finally we stack the groups for each
species together with np.concatenate (like pd.concat but works on numpy objects
and np.random.multivariate_normal returns numpy arrays not data frames) and put all of that in a
DataFrame using the feature names as the columns.
Then we add a species column, by repeating each species 20 times
[c]*N for c in gnb.classes_ and then unpack that into a single list instead of
as list of lists.
N = 20
gnb_df = pd.DataFrame(np.concatenate([np.random.multivariate_normal(th, sig*np.eye(4),N)
for th, sig in zip(gnb.theta_,gnb.sigma_)]),
columns = gnb.feature_names_in_)
gnb_df['species'] = [ci for cl in [[c]*N for c in gnb.classes_] for ci in cl]
sns.pairplot(data =gnb_df, hue='species')
/opt/hostedtoolcache/Python/3.8.13/x64/lib/python3.8/site-packages/sklearn/utils/deprecation.py:103: FutureWarning: Attribute `sigma_` was deprecated in 1.0 and will be removed in1.2. Use `var_` instead.
warnings.warn(msg, category=FutureWarning)
<seaborn.axisgrid.PairGrid at 0x7fed24f37e50>
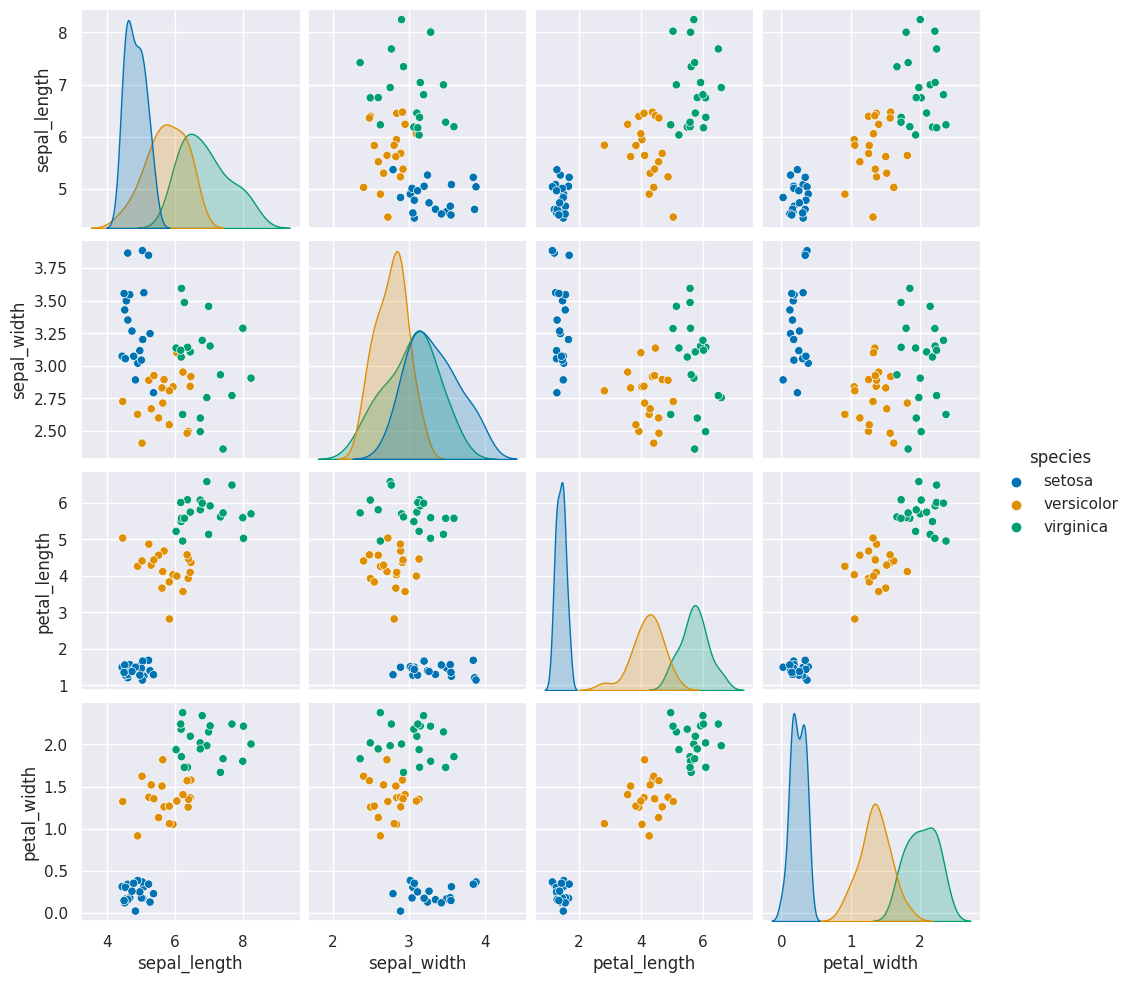
This one looks pretty close to the actual data. The biggest difference is that
these data are all in uniformly circular-ish blobs and the ones above are not.
That means that the naive assumption doesn’t hold perfectly on this data.
16.2. Evaluating#
On Wedneday, we used predict and checked the results
y_pred = gnb.predict(X_test)
y_pred
array(['virginica', 'versicolor', 'setosa', 'virginica', 'setosa',
'virginica', 'setosa', 'versicolor', 'versicolor', 'versicolor',
'versicolor', 'versicolor', 'versicolor', 'versicolor',
'versicolor', 'setosa', 'versicolor', 'versicolor', 'setosa',
'setosa', 'virginica', 'versicolor', 'setosa', 'setosa',
'virginica', 'setosa', 'setosa', 'versicolor', 'versicolor',
'setosa', 'virginica', 'versicolor', 'setosa', 'virginica',
'virginica', 'versicolor', 'setosa', 'versicolor', 'versicolor',
'versicolor', 'virginica', 'setosa', 'virginica', 'setosa',
'setosa', 'versicolor', 'virginica', 'virginica', 'versicolor',
'virginica', 'versicolor', 'virginica', 'versicolor', 'versicolor',
'virginica', 'versicolor', 'versicolor', 'virginica', 'versicolor',
'virginica', 'versicolor', 'setosa', 'virginica', 'versicolor',
'versicolor', 'versicolor', 'versicolor', 'virginica', 'setosa',
'setosa', 'virginica', 'versicolor', 'setosa', 'setosa',
'versicolor'], dtype='<U10')
Then compared to see how many matched the ground truth.
sum(y_pred == y_test)
71
out of the total number
len(y_test)
75
Scikit Learn also provides a score method for all of the estimator object
(different models). For a classifier, it provides the accuracy (percent correct).
gnb.score(X_test,y_test)
0.9466666666666667
which we could calculate with:
sum(y_pred == y_test)/len(y_test)
0.9466666666666667
The confusion matrix counts the number for each class (in this case the species) that truly have that value adn that were predicted to have that value. The wikipedia article on confusion matrices also summarized all of the different metrics. Its written in terms of two, outcomes, but we have 3.
confusion_matrix(y_test, y_pred,labels = gnb.classes_)
array([[21, 0, 0],
[ 0, 30, 0],
[ 0, 4, 20]])
Try it yourself
Could you create a Data Frame with columns for predicted and true class, then get the count of how many samples were in each combination? Does it match the table.
We can also get a report with a few metrics.
Recall is the percent of each species that were predicted correctly.
Precision is the percent of the ones predicted to be in a species that are truly that species.
the F1 score is combination of the two
print(classification_report(y_test,y_pred))
precision recall f1-score support
setosa 1.00 1.00 1.00 21
versicolor 0.88 1.00 0.94 30
virginica 1.00 0.83 0.91 24
accuracy 0.95 75
macro avg 0.96 0.94 0.95 75
weighted avg 0.95 0.95 0.95 75
16.3. Questions#
Ram token Opportunity
Contribute a question as an issue or contribute a soultion to one of the try it yourself above.