Clustering
Contents
23. Clustering#
Clustering is unsupervised learning. That means we do not have the labels to learn from. We aim to learn both the labels for each point and some way of characterizing the classes at the same time.
Computationally, this is a harder problem. Mathematically, we can typically solve problems when we have a number of equations equal to or greater than the number of unknowns. For \(N\) data points ind \(d\) dimensions and \(K\) clusters, we have \(N\) equations and \(N + K*d\) unknowns. This means we have a harder problem to solve.
For today, we’ll see K-means clustering which is defined by \(K\) a number of clusters and a mean (center) for each one. There are other K-centers algorithms for other types of centers.
Clustering is a stochastic (random) algorithm, so it can be a little harder to debug the models and measure performance. For this reason, we are going to lootk a little more closely at what it actuall does than we have with classification or regression.
import matplotlib.pyplot as plt
import numpy as np
import itertools as it
import seaborn as sns
from sklearn import datasets
from sklearn.cluster import KMeans
import string
import pandas as pd
23.1. How does Kmeans work?#
We will start with some synthetics data and then see how the clustering works.
C = 4
N = 200
offset = 2
spacing = 2
# choose the first C uppcase letters
classes = list(string.ascii_uppercase[:C])
# get the number of grid locations needed
G = int(np.ceil(np.sqrt(C)))
# get the locations for each axis
grid_locs = a = np.linspace(offset,offset+G*spacing,G)
# compute grid (i,j) for each combination of values above & keep C values
means = [(i,j) for i, j in it.product(grid_locs,grid_locs)][:C]
# store in dictionary with class labels
mu = {c: i for c, i in zip(classes,means)}
# random variances
sigma = {c: i*.5 for c, i in zip(classes,np.random.random(4))}
#randomly choose a class for each point, with equal probability
clusters_true = np.random.choice(classes,N)
# draw a randome point according to the means from above for each point
data = [np.random.multivariate_normal(mu[c],.25*np.eye(2)) for c in clusters_true]
# rounding to make display neater later
df = pd.DataFrame(data = data,columns = ['x' + str(i) for i in range(2)]).round(2)
# store in dataFram
df['true_cluster'] = clusters_true
sns.pairplot(data =df, hue='true_cluster')
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[2], line 29
26 # store in dataFram
27 df['true_cluster'] = clusters_true
---> 29 sns.pairplot(data =df, hue='true_cluster')
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/axisgrid.py:2148, in pairplot(data, hue, hue_order, palette, vars, x_vars, y_vars, kind, diag_kind, markers, height, aspect, corner, dropna, plot_kws, diag_kws, grid_kws, size)
2146 diag_kws.setdefault("fill", True)
2147 diag_kws.setdefault("warn_singular", False)
-> 2148 grid.map_diag(kdeplot, **diag_kws)
2150 # Maybe plot on the off-diagonals
2151 if diag_kind is not None:
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/axisgrid.py:1507, in PairGrid.map_diag(self, func, **kwargs)
1505 plot_kwargs.setdefault("hue_order", self._hue_order)
1506 plot_kwargs.setdefault("palette", self._orig_palette)
-> 1507 func(x=vector, **plot_kwargs)
1508 ax.legend_ = None
1510 self._add_axis_labels()
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/distributions.py:1717, in kdeplot(data, x, y, hue, weights, palette, hue_order, hue_norm, color, fill, multiple, common_norm, common_grid, cumulative, bw_method, bw_adjust, warn_singular, log_scale, levels, thresh, gridsize, cut, clip, legend, cbar, cbar_ax, cbar_kws, ax, **kwargs)
1713 if p.univariate:
1715 plot_kws = kwargs.copy()
-> 1717 p.plot_univariate_density(
1718 multiple=multiple,
1719 common_norm=common_norm,
1720 common_grid=common_grid,
1721 fill=fill,
1722 color=color,
1723 legend=legend,
1724 warn_singular=warn_singular,
1725 estimate_kws=estimate_kws,
1726 **plot_kws,
1727 )
1729 else:
1731 p.plot_bivariate_density(
1732 common_norm=common_norm,
1733 fill=fill,
(...)
1743 **kwargs,
1744 )
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/distributions.py:996, in _DistributionPlotter.plot_univariate_density(self, multiple, common_norm, common_grid, warn_singular, fill, color, legend, estimate_kws, **plot_kws)
993 if "x" in self.variables:
995 if fill:
--> 996 artist = ax.fill_between(support, fill_from, density, **artist_kws)
998 else:
999 artist, = ax.plot(support, density, **artist_kws)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/matplotlib/__init__.py:1423, in _preprocess_data.<locals>.inner(ax, data, *args, **kwargs)
1420 @functools.wraps(func)
1421 def inner(ax, *args, data=None, **kwargs):
1422 if data is None:
-> 1423 return func(ax, *map(sanitize_sequence, args), **kwargs)
1425 bound = new_sig.bind(ax, *args, **kwargs)
1426 auto_label = (bound.arguments.get(label_namer)
1427 or bound.kwargs.get(label_namer))
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/matplotlib/axes/_axes.py:5367, in Axes.fill_between(self, x, y1, y2, where, interpolate, step, **kwargs)
5365 def fill_between(self, x, y1, y2=0, where=None, interpolate=False,
5366 step=None, **kwargs):
-> 5367 return self._fill_between_x_or_y(
5368 "x", x, y1, y2,
5369 where=where, interpolate=interpolate, step=step, **kwargs)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/matplotlib/axes/_axes.py:5272, in Axes._fill_between_x_or_y(self, ind_dir, ind, dep1, dep2, where, interpolate, step, **kwargs)
5268 kwargs["facecolor"] = \
5269 self._get_patches_for_fill.get_next_color()
5271 # Handle united data, such as dates
-> 5272 ind, dep1, dep2 = map(
5273 ma.masked_invalid, self._process_unit_info(
5274 [(ind_dir, ind), (dep_dir, dep1), (dep_dir, dep2)], kwargs))
5276 for name, array in [
5277 (ind_dir, ind), (f"{dep_dir}1", dep1), (f"{dep_dir}2", dep2)]:
5278 if array.ndim > 1:
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/numpy/ma/core.py:2360, in masked_invalid(a, copy)
2332 def masked_invalid(a, copy=True):
2333 """
2334 Mask an array where invalid values occur (NaNs or infs).
2335
(...)
2357
2358 """
-> 2360 return masked_where(~(np.isfinite(getdata(a))), a, copy=copy)
TypeError: ufunc 'isfinite' not supported for the input types, and the inputs could not be safely coerced to any supported types according to the casting rule ''safe''
df.head()
| x0 | x1 | true_cluster | |
|---|---|---|---|
| 0 | 2.94 | 6.45 | B |
| 1 | 6.08 | 2.08 | C |
| 2 | 2.04 | 6.30 | B |
| 3 | 1.49 | 2.37 | A |
| 4 | 6.02 | 5.20 | D |
23.2. Kmeans#
Next, we’ll pick 4 random points to be the starting points as the means.
K = 4
mu0 = df.sample(n=K).values
mu0
array([[5.72, 1.01, 'C'],
[1.88, 6.52, 'B'],
[1.56, 5.73, 'B'],
[1.57, 5.51, 'B']], dtype=object)
Now, we will compute, fo each sample which of those four points it is closest to first by taking the difference, squaring it, then summing along each row.
[((df-mu_i)**2).sum(axis=1) for mu_i in mu0]
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:165, in _na_arithmetic_op(left, right, op, is_cmp)
164 try:
--> 165 result = func(left, right)
166 except TypeError:
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:241, in evaluate(op, a, b, use_numexpr)
239 if use_numexpr:
240 # error: "None" not callable
--> 241 return _evaluate(op, op_str, a, b) # type: ignore[misc]
242 return _evaluate_standard(op, op_str, a, b)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:70, in _evaluate_standard(op, op_str, a, b)
69 _store_test_result(False)
---> 70 return op(a, b)
TypeError: unsupported operand type(s) for -: 'str' and 'str'
During handling of the above exception, another exception occurred:
TypeError Traceback (most recent call last)
Cell In[5], line 1
----> 1 [((df-mu_i)**2).sum(axis=1) for mu_i in mu0]
Cell In[5], line 1, in <listcomp>(.0)
----> 1 [((df-mu_i)**2).sum(axis=1) for mu_i in mu0]
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/common.py:72, in _unpack_zerodim_and_defer.<locals>.new_method(self, other)
68 return NotImplemented
70 other = item_from_zerodim(other)
---> 72 return method(self, other)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/arraylike.py:110, in OpsMixin.__sub__(self, other)
108 @unpack_zerodim_and_defer("__sub__")
109 def __sub__(self, other):
--> 110 return self._arith_method(other, operator.sub)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7591, in DataFrame._arith_method(self, other, op)
7587 other = ops.maybe_prepare_scalar_for_op(other, (self.shape[axis],))
7589 self, other = ops.align_method_FRAME(self, other, axis, flex=True, level=None)
-> 7591 new_data = self._dispatch_frame_op(other, op, axis=axis)
7592 return self._construct_result(new_data)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7630, in DataFrame._dispatch_frame_op(self, right, func, axis)
7624 # TODO: The previous assertion `assert right._indexed_same(self)`
7625 # fails in cases with empty columns reached via
7626 # _frame_arith_method_with_reindex
7627
7628 # TODO operate_blockwise expects a manager of the same type
7629 with np.errstate(all="ignore"):
-> 7630 bm = self._mgr.operate_blockwise(
7631 # error: Argument 1 to "operate_blockwise" of "ArrayManager" has
7632 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7633 # "ArrayManager"
7634 # error: Argument 1 to "operate_blockwise" of "BlockManager" has
7635 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7636 # "BlockManager"
7637 right._mgr, # type: ignore[arg-type]
7638 array_op,
7639 )
7640 return self._constructor(bm)
7642 elif isinstance(right, Series) and axis == 1:
7643 # axis=1 means we want to operate row-by-row
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/managers.py:1586, in BlockManager.operate_blockwise(self, other, array_op)
1582 def operate_blockwise(self, other: BlockManager, array_op) -> BlockManager:
1583 """
1584 Apply array_op blockwise with another (aligned) BlockManager.
1585 """
-> 1586 return operate_blockwise(self, other, array_op)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/ops.py:63, in operate_blockwise(left, right, array_op)
61 res_blks: list[Block] = []
62 for lvals, rvals, locs, left_ea, right_ea, rblk in _iter_block_pairs(left, right):
---> 63 res_values = array_op(lvals, rvals)
64 if left_ea and not right_ea and hasattr(res_values, "reshape"):
65 res_values = res_values.reshape(1, -1)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:226, in arithmetic_op(left, right, op)
222 _bool_arith_check(op, left, right)
224 # error: Argument 1 to "_na_arithmetic_op" has incompatible type
225 # "Union[ExtensionArray, ndarray[Any, Any]]"; expected "ndarray[Any, Any]"
--> 226 res_values = _na_arithmetic_op(left, right, op) # type: ignore[arg-type]
228 return res_values
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:172, in _na_arithmetic_op(left, right, op, is_cmp)
166 except TypeError:
167 if not is_cmp and (is_object_dtype(left.dtype) or is_object_dtype(right)):
168 # For object dtype, fallback to a masked operation (only operating
169 # on the non-missing values)
170 # Don't do this for comparisons, as that will handle complex numbers
171 # incorrectly, see GH#32047
--> 172 result = _masked_arith_op(left, right, op)
173 else:
174 raise
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:110, in _masked_arith_op(x, y, op)
108 # See GH#5284, GH#5035, GH#19448 for historical reference
109 if mask.any():
--> 110 result[mask] = op(xrav[mask], yrav[mask])
112 else:
113 if not is_scalar(y):
TypeError: unsupported operand type(s) for -: 'str' and 'str'
This gives us a list of 4 data DataFrames, one for each mean (mu), with one row for each point in the dataset with the distance from that point to the corresponding mean. We can stack these into one DataFrame.
pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).head()
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:165, in _na_arithmetic_op(left, right, op, is_cmp)
164 try:
--> 165 result = func(left, right)
166 except TypeError:
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:241, in evaluate(op, a, b, use_numexpr)
239 if use_numexpr:
240 # error: "None" not callable
--> 241 return _evaluate(op, op_str, a, b) # type: ignore[misc]
242 return _evaluate_standard(op, op_str, a, b)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:70, in _evaluate_standard(op, op_str, a, b)
69 _store_test_result(False)
---> 70 return op(a, b)
TypeError: unsupported operand type(s) for -: 'str' and 'str'
During handling of the above exception, another exception occurred:
TypeError Traceback (most recent call last)
Cell In[6], line 1
----> 1 pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).head()
Cell In[6], line 1, in <listcomp>(.0)
----> 1 pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).head()
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/common.py:72, in _unpack_zerodim_and_defer.<locals>.new_method(self, other)
68 return NotImplemented
70 other = item_from_zerodim(other)
---> 72 return method(self, other)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/arraylike.py:110, in OpsMixin.__sub__(self, other)
108 @unpack_zerodim_and_defer("__sub__")
109 def __sub__(self, other):
--> 110 return self._arith_method(other, operator.sub)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7591, in DataFrame._arith_method(self, other, op)
7587 other = ops.maybe_prepare_scalar_for_op(other, (self.shape[axis],))
7589 self, other = ops.align_method_FRAME(self, other, axis, flex=True, level=None)
-> 7591 new_data = self._dispatch_frame_op(other, op, axis=axis)
7592 return self._construct_result(new_data)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7630, in DataFrame._dispatch_frame_op(self, right, func, axis)
7624 # TODO: The previous assertion `assert right._indexed_same(self)`
7625 # fails in cases with empty columns reached via
7626 # _frame_arith_method_with_reindex
7627
7628 # TODO operate_blockwise expects a manager of the same type
7629 with np.errstate(all="ignore"):
-> 7630 bm = self._mgr.operate_blockwise(
7631 # error: Argument 1 to "operate_blockwise" of "ArrayManager" has
7632 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7633 # "ArrayManager"
7634 # error: Argument 1 to "operate_blockwise" of "BlockManager" has
7635 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7636 # "BlockManager"
7637 right._mgr, # type: ignore[arg-type]
7638 array_op,
7639 )
7640 return self._constructor(bm)
7642 elif isinstance(right, Series) and axis == 1:
7643 # axis=1 means we want to operate row-by-row
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/managers.py:1586, in BlockManager.operate_blockwise(self, other, array_op)
1582 def operate_blockwise(self, other: BlockManager, array_op) -> BlockManager:
1583 """
1584 Apply array_op blockwise with another (aligned) BlockManager.
1585 """
-> 1586 return operate_blockwise(self, other, array_op)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/ops.py:63, in operate_blockwise(left, right, array_op)
61 res_blks: list[Block] = []
62 for lvals, rvals, locs, left_ea, right_ea, rblk in _iter_block_pairs(left, right):
---> 63 res_values = array_op(lvals, rvals)
64 if left_ea and not right_ea and hasattr(res_values, "reshape"):
65 res_values = res_values.reshape(1, -1)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:226, in arithmetic_op(left, right, op)
222 _bool_arith_check(op, left, right)
224 # error: Argument 1 to "_na_arithmetic_op" has incompatible type
225 # "Union[ExtensionArray, ndarray[Any, Any]]"; expected "ndarray[Any, Any]"
--> 226 res_values = _na_arithmetic_op(left, right, op) # type: ignore[arg-type]
228 return res_values
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:172, in _na_arithmetic_op(left, right, op, is_cmp)
166 except TypeError:
167 if not is_cmp and (is_object_dtype(left.dtype) or is_object_dtype(right)):
168 # For object dtype, fallback to a masked operation (only operating
169 # on the non-missing values)
170 # Don't do this for comparisons, as that will handle complex numbers
171 # incorrectly, see GH#32047
--> 172 result = _masked_arith_op(left, right, op)
173 else:
174 raise
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:110, in _masked_arith_op(x, y, op)
108 # See GH#5284, GH#5035, GH#19448 for historical reference
109 if mask.any():
--> 110 result[mask] = op(xrav[mask], yrav[mask])
112 else:
113 if not is_scalar(y):
TypeError: unsupported operand type(s) for -: 'str' and 'str'
Now we have one row per sample and one column per mean, with with the distance from that point to the mean. What we want is to calculate the assignment, which
mean is closest, for each point. Using idxmin with axis=1 we take the
minimum across each row and returns the index (location) of that minimum.
pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).idxmin(axis=1).head()
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:165, in _na_arithmetic_op(left, right, op, is_cmp)
164 try:
--> 165 result = func(left, right)
166 except TypeError:
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:241, in evaluate(op, a, b, use_numexpr)
239 if use_numexpr:
240 # error: "None" not callable
--> 241 return _evaluate(op, op_str, a, b) # type: ignore[misc]
242 return _evaluate_standard(op, op_str, a, b)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:70, in _evaluate_standard(op, op_str, a, b)
69 _store_test_result(False)
---> 70 return op(a, b)
TypeError: unsupported operand type(s) for -: 'str' and 'str'
During handling of the above exception, another exception occurred:
TypeError Traceback (most recent call last)
Cell In[7], line 1
----> 1 pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).idxmin(axis=1).head()
Cell In[7], line 1, in <listcomp>(.0)
----> 1 pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).idxmin(axis=1).head()
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/common.py:72, in _unpack_zerodim_and_defer.<locals>.new_method(self, other)
68 return NotImplemented
70 other = item_from_zerodim(other)
---> 72 return method(self, other)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/arraylike.py:110, in OpsMixin.__sub__(self, other)
108 @unpack_zerodim_and_defer("__sub__")
109 def __sub__(self, other):
--> 110 return self._arith_method(other, operator.sub)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7591, in DataFrame._arith_method(self, other, op)
7587 other = ops.maybe_prepare_scalar_for_op(other, (self.shape[axis],))
7589 self, other = ops.align_method_FRAME(self, other, axis, flex=True, level=None)
-> 7591 new_data = self._dispatch_frame_op(other, op, axis=axis)
7592 return self._construct_result(new_data)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7630, in DataFrame._dispatch_frame_op(self, right, func, axis)
7624 # TODO: The previous assertion `assert right._indexed_same(self)`
7625 # fails in cases with empty columns reached via
7626 # _frame_arith_method_with_reindex
7627
7628 # TODO operate_blockwise expects a manager of the same type
7629 with np.errstate(all="ignore"):
-> 7630 bm = self._mgr.operate_blockwise(
7631 # error: Argument 1 to "operate_blockwise" of "ArrayManager" has
7632 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7633 # "ArrayManager"
7634 # error: Argument 1 to "operate_blockwise" of "BlockManager" has
7635 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7636 # "BlockManager"
7637 right._mgr, # type: ignore[arg-type]
7638 array_op,
7639 )
7640 return self._constructor(bm)
7642 elif isinstance(right, Series) and axis == 1:
7643 # axis=1 means we want to operate row-by-row
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/managers.py:1586, in BlockManager.operate_blockwise(self, other, array_op)
1582 def operate_blockwise(self, other: BlockManager, array_op) -> BlockManager:
1583 """
1584 Apply array_op blockwise with another (aligned) BlockManager.
1585 """
-> 1586 return operate_blockwise(self, other, array_op)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/ops.py:63, in operate_blockwise(left, right, array_op)
61 res_blks: list[Block] = []
62 for lvals, rvals, locs, left_ea, right_ea, rblk in _iter_block_pairs(left, right):
---> 63 res_values = array_op(lvals, rvals)
64 if left_ea and not right_ea and hasattr(res_values, "reshape"):
65 res_values = res_values.reshape(1, -1)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:226, in arithmetic_op(left, right, op)
222 _bool_arith_check(op, left, right)
224 # error: Argument 1 to "_na_arithmetic_op" has incompatible type
225 # "Union[ExtensionArray, ndarray[Any, Any]]"; expected "ndarray[Any, Any]"
--> 226 res_values = _na_arithmetic_op(left, right, op) # type: ignore[arg-type]
228 return res_values
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:172, in _na_arithmetic_op(left, right, op, is_cmp)
166 except TypeError:
167 if not is_cmp and (is_object_dtype(left.dtype) or is_object_dtype(right)):
168 # For object dtype, fallback to a masked operation (only operating
169 # on the non-missing values)
170 # Don't do this for comparisons, as that will handle complex numbers
171 # incorrectly, see GH#32047
--> 172 result = _masked_arith_op(left, right, op)
173 else:
174 raise
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:110, in _masked_arith_op(x, y, op)
108 # See GH#5284, GH#5035, GH#19448 for historical reference
109 if mask.any():
--> 110 result[mask] = op(xrav[mask], yrav[mask])
112 else:
113 if not is_scalar(y):
TypeError: unsupported operand type(s) for -: 'str' and 'str'
We’ll save all of this in a column named '0'. Since it is our 0th iteration.
df['0'] = pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).idxmin(axis=1)
df.head()
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:165, in _na_arithmetic_op(left, right, op, is_cmp)
164 try:
--> 165 result = func(left, right)
166 except TypeError:
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:241, in evaluate(op, a, b, use_numexpr)
239 if use_numexpr:
240 # error: "None" not callable
--> 241 return _evaluate(op, op_str, a, b) # type: ignore[misc]
242 return _evaluate_standard(op, op_str, a, b)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/computation/expressions.py:70, in _evaluate_standard(op, op_str, a, b)
69 _store_test_result(False)
---> 70 return op(a, b)
TypeError: unsupported operand type(s) for -: 'str' and 'str'
During handling of the above exception, another exception occurred:
TypeError Traceback (most recent call last)
Cell In[8], line 1
----> 1 df['0'] = pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).idxmin(axis=1)
3 df.head()
Cell In[8], line 1, in <listcomp>(.0)
----> 1 df['0'] = pd.concat([((df-mu_i)**2).sum(axis=1) for mu_i in mu0],axis=1).idxmin(axis=1)
3 df.head()
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/common.py:72, in _unpack_zerodim_and_defer.<locals>.new_method(self, other)
68 return NotImplemented
70 other = item_from_zerodim(other)
---> 72 return method(self, other)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/arraylike.py:110, in OpsMixin.__sub__(self, other)
108 @unpack_zerodim_and_defer("__sub__")
109 def __sub__(self, other):
--> 110 return self._arith_method(other, operator.sub)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7591, in DataFrame._arith_method(self, other, op)
7587 other = ops.maybe_prepare_scalar_for_op(other, (self.shape[axis],))
7589 self, other = ops.align_method_FRAME(self, other, axis, flex=True, level=None)
-> 7591 new_data = self._dispatch_frame_op(other, op, axis=axis)
7592 return self._construct_result(new_data)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:7630, in DataFrame._dispatch_frame_op(self, right, func, axis)
7624 # TODO: The previous assertion `assert right._indexed_same(self)`
7625 # fails in cases with empty columns reached via
7626 # _frame_arith_method_with_reindex
7627
7628 # TODO operate_blockwise expects a manager of the same type
7629 with np.errstate(all="ignore"):
-> 7630 bm = self._mgr.operate_blockwise(
7631 # error: Argument 1 to "operate_blockwise" of "ArrayManager" has
7632 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7633 # "ArrayManager"
7634 # error: Argument 1 to "operate_blockwise" of "BlockManager" has
7635 # incompatible type "Union[ArrayManager, BlockManager]"; expected
7636 # "BlockManager"
7637 right._mgr, # type: ignore[arg-type]
7638 array_op,
7639 )
7640 return self._constructor(bm)
7642 elif isinstance(right, Series) and axis == 1:
7643 # axis=1 means we want to operate row-by-row
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/managers.py:1586, in BlockManager.operate_blockwise(self, other, array_op)
1582 def operate_blockwise(self, other: BlockManager, array_op) -> BlockManager:
1583 """
1584 Apply array_op blockwise with another (aligned) BlockManager.
1585 """
-> 1586 return operate_blockwise(self, other, array_op)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/ops.py:63, in operate_blockwise(left, right, array_op)
61 res_blks: list[Block] = []
62 for lvals, rvals, locs, left_ea, right_ea, rblk in _iter_block_pairs(left, right):
---> 63 res_values = array_op(lvals, rvals)
64 if left_ea and not right_ea and hasattr(res_values, "reshape"):
65 res_values = res_values.reshape(1, -1)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:226, in arithmetic_op(left, right, op)
222 _bool_arith_check(op, left, right)
224 # error: Argument 1 to "_na_arithmetic_op" has incompatible type
225 # "Union[ExtensionArray, ndarray[Any, Any]]"; expected "ndarray[Any, Any]"
--> 226 res_values = _na_arithmetic_op(left, right, op) # type: ignore[arg-type]
228 return res_values
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:172, in _na_arithmetic_op(left, right, op, is_cmp)
166 except TypeError:
167 if not is_cmp and (is_object_dtype(left.dtype) or is_object_dtype(right)):
168 # For object dtype, fallback to a masked operation (only operating
169 # on the non-missing values)
170 # Don't do this for comparisons, as that will handle complex numbers
171 # incorrectly, see GH#32047
--> 172 result = _masked_arith_op(left, right, op)
173 else:
174 raise
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/ops/array_ops.py:110, in _masked_arith_op(x, y, op)
108 # See GH#5284, GH#5035, GH#19448 for historical reference
109 if mask.any():
--> 110 result[mask] = op(xrav[mask], yrav[mask])
112 else:
113 if not is_scalar(y):
TypeError: unsupported operand type(s) for -: 'str' and 'str'
Here, we’ll set up some helper code.
data_cols = ['x0','x1']
def mu_to_df(mu,i):
mu_df = pd.DataFrame(mu,columns=data_cols)
mu_df['iteration'] = str(i)
mu_df['class'] = ['M'+str(i) for i in range(K)]
mu_df['type'] = 'mu'
return mu_df
# color maps, we select every other value from this palette that has 8 values & is paird
cmap_pt = sns.color_palette('tab20',8)[1::2] # starting from 1
cmap_mu = sns.color_palette('tab20',8)[0::2] # starting form 0
Now we can plot the data, save the axis, and plot the means on top of that.
Seaborn plotting functions return an axis, by saving that to a variable, we
can pass it to the ax parameter of another plotting function so that both
plotting functions go on the same figure.
sfig = sns.scatterplot(data=df,x='x0',y='x1', hue='0',
palette =cmap_pt,legend=False)
mu_df = mu_to_df(mu0,0)
sns.scatterplot(data=mu_df, x='x0',y='x1',hue='class',
palette=cmap_mu,legend=False, ax=sfig)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[10], line 1
----> 1 sfig = sns.scatterplot(data=df,x='x0',y='x1', hue='0',
2 palette =cmap_pt,legend=False)
3 mu_df = mu_to_df(mu0,0)
4 sns.scatterplot(data=mu_df, x='x0',y='x1',hue='class',
5 palette=cmap_mu,legend=False, ax=sfig)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/relational.py:742, in scatterplot(data, x, y, hue, size, style, palette, hue_order, hue_norm, sizes, size_order, size_norm, markers, style_order, legend, ax, **kwargs)
732 def scatterplot(
733 data=None, *,
734 x=None, y=None, hue=None, size=None, style=None,
(...)
738 **kwargs
739 ):
741 variables = _ScatterPlotter.get_semantics(locals())
--> 742 p = _ScatterPlotter(data=data, variables=variables, legend=legend)
744 p.map_hue(palette=palette, order=hue_order, norm=hue_norm)
745 p.map_size(sizes=sizes, order=size_order, norm=size_norm)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/relational.py:538, in _ScatterPlotter.__init__(self, data, variables, legend)
529 def __init__(self, *, data=None, variables={}, legend=None):
530
531 # TODO this is messy, we want the mapping to be agnostic about
532 # the kind of plot to draw, but for the time being we need to set
533 # this information so the SizeMapping can use it
534 self._default_size_range = (
535 np.r_[.5, 2] * np.square(mpl.rcParams["lines.markersize"])
536 )
--> 538 super().__init__(data=data, variables=variables)
540 self.legend = legend
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:640, in VectorPlotter.__init__(self, data, variables)
635 # var_ordered is relevant only for categorical axis variables, and may
636 # be better handled by an internal axis information object that tracks
637 # such information and is set up by the scale_* methods. The analogous
638 # information for numeric axes would be information about log scales.
639 self._var_ordered = {"x": False, "y": False} # alt., used DefaultDict
--> 640 self.assign_variables(data, variables)
642 for var, cls in self._semantic_mappings.items():
643
644 # Create the mapping function
645 map_func = partial(cls.map, plotter=self)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:701, in VectorPlotter.assign_variables(self, data, variables)
699 else:
700 self.input_format = "long"
--> 701 plot_data, variables = self._assign_variables_longform(
702 data, **variables,
703 )
705 self.plot_data = plot_data
706 self.variables = variables
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:938, in VectorPlotter._assign_variables_longform(self, data, **kwargs)
933 elif isinstance(val, (str, bytes)):
934
935 # This looks like a column name but we don't know what it means!
937 err = f"Could not interpret value `{val}` for parameter `{key}`"
--> 938 raise ValueError(err)
940 else:
941
942 # Otherwise, assume the value is itself data
943
944 # Raise when data object is present and a vector can't matched
945 if isinstance(data, pd.DataFrame) and not isinstance(val, pd.Series):
ValueError: Could not interpret value `0` for parameter `hue`
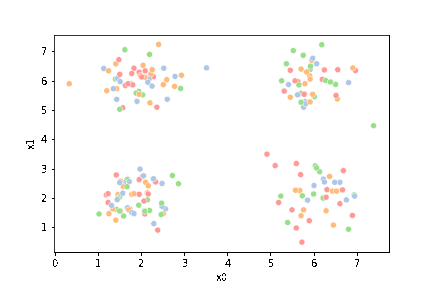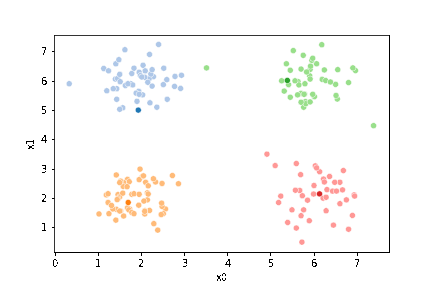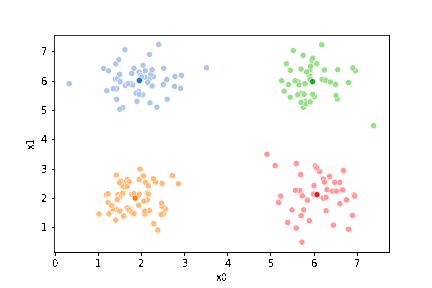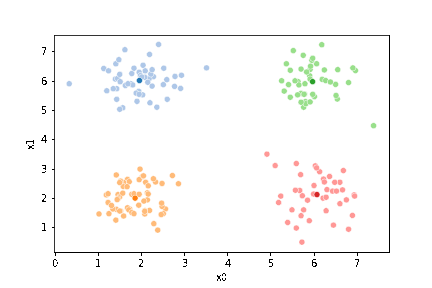
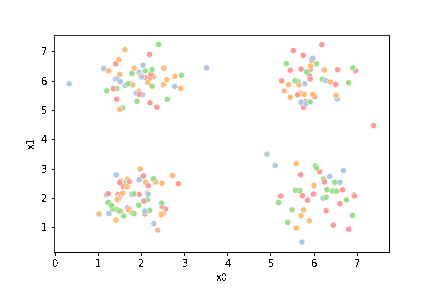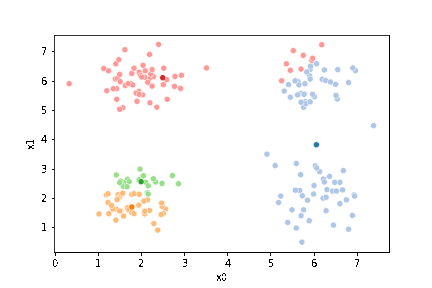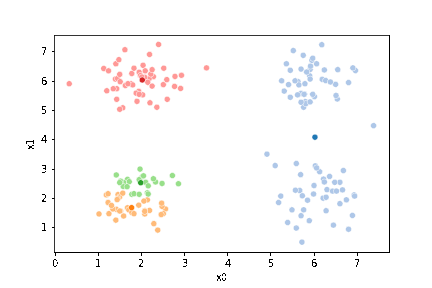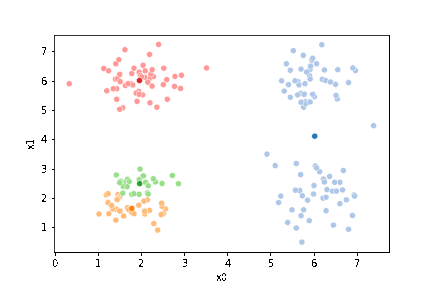
We see that each point is assigned to the lighter shade of its matching mean. These points are the one that is closest to each point, but they’re not the centers of the point clouds. Now, we can compute new means of the points assigned to each cluster, using groupby.
mu1 = df.groupby('0')[data_cols].mean().values
mu1
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[11], line 1
----> 1 mu1 = df.groupby('0')[data_cols].mean().values
2 mu1
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:8399, in DataFrame.groupby(self, by, axis, level, as_index, sort, group_keys, squeeze, observed, dropna)
8396 raise TypeError("You have to supply one of 'by' and 'level'")
8397 axis = self._get_axis_number(axis)
-> 8399 return DataFrameGroupBy(
8400 obj=self,
8401 keys=by,
8402 axis=axis,
8403 level=level,
8404 as_index=as_index,
8405 sort=sort,
8406 group_keys=group_keys,
8407 squeeze=squeeze,
8408 observed=observed,
8409 dropna=dropna,
8410 )
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/groupby/groupby.py:959, in GroupBy.__init__(self, obj, keys, axis, level, grouper, exclusions, selection, as_index, sort, group_keys, squeeze, observed, mutated, dropna)
956 if grouper is None:
957 from pandas.core.groupby.grouper import get_grouper
--> 959 grouper, exclusions, obj = get_grouper(
960 obj,
961 keys,
962 axis=axis,
963 level=level,
964 sort=sort,
965 observed=observed,
966 mutated=self.mutated,
967 dropna=self.dropna,
968 )
970 self.obj = obj
971 self.axis = obj._get_axis_number(axis)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/groupby/grouper.py:888, in get_grouper(obj, key, axis, level, sort, observed, mutated, validate, dropna)
886 in_axis, level, gpr = False, gpr, None
887 else:
--> 888 raise KeyError(gpr)
889 elif isinstance(gpr, Grouper) and gpr.key is not None:
890 # Add key to exclusions
891 exclusions.add(gpr.key)
KeyError: '0'
We can plot these again, the same data, but with the new means.
sfig = sns.scatterplot(data=df,x='x0',y='x1', hue='0',
palette =cmap_pt,legend=False)
mu_df = mu_to_df(mu1,0)
sns.scatterplot(data=mu_df, x='x0',y='x1',hue='class',
palette=cmap_mu,legend=False, ax=sfig)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[12], line 1
----> 1 sfig = sns.scatterplot(data=df,x='x0',y='x1', hue='0',
2 palette =cmap_pt,legend=False)
3 mu_df = mu_to_df(mu1,0)
4 sns.scatterplot(data=mu_df, x='x0',y='x1',hue='class',
5 palette=cmap_mu,legend=False, ax=sfig)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/relational.py:742, in scatterplot(data, x, y, hue, size, style, palette, hue_order, hue_norm, sizes, size_order, size_norm, markers, style_order, legend, ax, **kwargs)
732 def scatterplot(
733 data=None, *,
734 x=None, y=None, hue=None, size=None, style=None,
(...)
738 **kwargs
739 ):
741 variables = _ScatterPlotter.get_semantics(locals())
--> 742 p = _ScatterPlotter(data=data, variables=variables, legend=legend)
744 p.map_hue(palette=palette, order=hue_order, norm=hue_norm)
745 p.map_size(sizes=sizes, order=size_order, norm=size_norm)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/relational.py:538, in _ScatterPlotter.__init__(self, data, variables, legend)
529 def __init__(self, *, data=None, variables={}, legend=None):
530
531 # TODO this is messy, we want the mapping to be agnostic about
532 # the kind of plot to draw, but for the time being we need to set
533 # this information so the SizeMapping can use it
534 self._default_size_range = (
535 np.r_[.5, 2] * np.square(mpl.rcParams["lines.markersize"])
536 )
--> 538 super().__init__(data=data, variables=variables)
540 self.legend = legend
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:640, in VectorPlotter.__init__(self, data, variables)
635 # var_ordered is relevant only for categorical axis variables, and may
636 # be better handled by an internal axis information object that tracks
637 # such information and is set up by the scale_* methods. The analogous
638 # information for numeric axes would be information about log scales.
639 self._var_ordered = {"x": False, "y": False} # alt., used DefaultDict
--> 640 self.assign_variables(data, variables)
642 for var, cls in self._semantic_mappings.items():
643
644 # Create the mapping function
645 map_func = partial(cls.map, plotter=self)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:701, in VectorPlotter.assign_variables(self, data, variables)
699 else:
700 self.input_format = "long"
--> 701 plot_data, variables = self._assign_variables_longform(
702 data, **variables,
703 )
705 self.plot_data = plot_data
706 self.variables = variables
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:938, in VectorPlotter._assign_variables_longform(self, data, **kwargs)
933 elif isinstance(val, (str, bytes)):
934
935 # This looks like a column name but we don't know what it means!
937 err = f"Could not interpret value `{val}` for parameter `{key}`"
--> 938 raise ValueError(err)
940 else:
941
942 # Otherwise, assume the value is itself data
943
944 # Raise when data object is present and a vector can't matched
945 if isinstance(data, pd.DataFrame) and not isinstance(val, pd.Series):
ValueError: Could not interpret value `0` for parameter `hue`
We see that now the means are in the center of each cluster, but that there are now points in one color that are assigned to other clusters.
So, again we can update the assignments.
df['1'] = pd.concat([((df[data_cols]-mu_i)**2).sum(axis=1) for mu_i in mu1],axis=1).idxmin(axis=1)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[13], line 1
----> 1 df['1'] = pd.concat([((df[data_cols]-mu_i)**2).sum(axis=1) for mu_i in mu1],axis=1).idxmin(axis=1)
NameError: name 'mu1' is not defined
And plot again.
sfig = sns.scatterplot(data=df,x='x0',y='x1', hue='0',
palette =cmap_pt,legend=False)
mu_df = mu_to_df(mu,0)
sns.scatterplot(data=mu_df, x='x0',y='x1',hue='class',
palette=cmap_mu,legend=False, ax=sfig)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[14], line 1
----> 1 sfig = sns.scatterplot(data=df,x='x0',y='x1', hue='0',
2 palette =cmap_pt,legend=False)
3 mu_df = mu_to_df(mu,0)
4 sns.scatterplot(data=mu_df, x='x0',y='x1',hue='class',
5 palette=cmap_mu,legend=False, ax=sfig)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/relational.py:742, in scatterplot(data, x, y, hue, size, style, palette, hue_order, hue_norm, sizes, size_order, size_norm, markers, style_order, legend, ax, **kwargs)
732 def scatterplot(
733 data=None, *,
734 x=None, y=None, hue=None, size=None, style=None,
(...)
738 **kwargs
739 ):
741 variables = _ScatterPlotter.get_semantics(locals())
--> 742 p = _ScatterPlotter(data=data, variables=variables, legend=legend)
744 p.map_hue(palette=palette, order=hue_order, norm=hue_norm)
745 p.map_size(sizes=sizes, order=size_order, norm=size_norm)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/relational.py:538, in _ScatterPlotter.__init__(self, data, variables, legend)
529 def __init__(self, *, data=None, variables={}, legend=None):
530
531 # TODO this is messy, we want the mapping to be agnostic about
532 # the kind of plot to draw, but for the time being we need to set
533 # this information so the SizeMapping can use it
534 self._default_size_range = (
535 np.r_[.5, 2] * np.square(mpl.rcParams["lines.markersize"])
536 )
--> 538 super().__init__(data=data, variables=variables)
540 self.legend = legend
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:640, in VectorPlotter.__init__(self, data, variables)
635 # var_ordered is relevant only for categorical axis variables, and may
636 # be better handled by an internal axis information object that tracks
637 # such information and is set up by the scale_* methods. The analogous
638 # information for numeric axes would be information about log scales.
639 self._var_ordered = {"x": False, "y": False} # alt., used DefaultDict
--> 640 self.assign_variables(data, variables)
642 for var, cls in self._semantic_mappings.items():
643
644 # Create the mapping function
645 map_func = partial(cls.map, plotter=self)
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:701, in VectorPlotter.assign_variables(self, data, variables)
699 else:
700 self.input_format = "long"
--> 701 plot_data, variables = self._assign_variables_longform(
702 data, **variables,
703 )
705 self.plot_data = plot_data
706 self.variables = variables
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/seaborn/_oldcore.py:938, in VectorPlotter._assign_variables_longform(self, data, **kwargs)
933 elif isinstance(val, (str, bytes)):
934
935 # This looks like a column name but we don't know what it means!
937 err = f"Could not interpret value `{val}` for parameter `{key}`"
--> 938 raise ValueError(err)
940 else:
941
942 # Otherwise, assume the value is itself data
943
944 # Raise when data object is present and a vector can't matched
945 if isinstance(data, pd.DataFrame) and not isinstance(val, pd.Series):
ValueError: Could not interpret value `0` for parameter `hue`
If we keep going back and forth like this, eventually, the assignment step will not change any assignments. We call this condition convergence. We can implement the algorithm with a while loop.
Correction
In the following I swapped the order of the mean update and assignment steps.
My previous version had a different initialization (the above part) so it
was okay for the steps to be in the other order.
i =1
mu_list = [mu_to_df(mu0,i), mu_to_df(mu1,i)]
cur_old = str(i-1)
cur_new = str(i)
while sum(df[cur_old] !=df[cur_new]) >0:
cur_old = cur_new
i +=1
cur_new = str(i)
# update the means and plot with current generating assignments
mu = df.groupby(cur_old)[data_cols].mean().values
mu_df = mu_to_df(mu,i)
mu_list.append(mu_df)
fig = plt.figure()
sfig = sns.scatterplot(data =df,x='x0',y='x1',hue=cur_old,palette=cmap_pt,legend=False)
sns.scatterplot(data =mu_df,x='x0',y='x1',hue='class',palette=cmap_mu,ax=sfig,legend=False)
# update the assigments and plot with the associated means
df[cur_new] = pd.concat([((df[data_cols]-mu_i)**2).sum(axis=1) for mu_i in mu],axis=1).idxmin(axis=1)
fig = plt.figure()
sfig = sns.scatterplot(data =df,x='x0',y='x1',hue=cur_new,palette=cmap_pt,legend=False)
sns.scatterplot(data =mu_df,x='x0',y='x1',hue='class',palette=cmap_mu,ax=sfig,legend=False)
n_iter = i
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[15], line 2
1 i =1
----> 2 mu_list = [mu_to_df(mu0,i), mu_to_df(mu1,i)]
3 cur_old = str(i-1)
4 cur_new = str(i)
Cell In[9], line 3, in mu_to_df(mu, i)
2 def mu_to_df(mu,i):
----> 3 mu_df = pd.DataFrame(mu,columns=data_cols)
4 mu_df['iteration'] = str(i)
5 mu_df['class'] = ['M'+str(i) for i in range(K)]
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/frame.py:721, in DataFrame.__init__(self, data, index, columns, dtype, copy)
711 mgr = dict_to_mgr(
712 # error: Item "ndarray" of "Union[ndarray, Series, Index]" has no
713 # attribute "name"
(...)
718 typ=manager,
719 )
720 else:
--> 721 mgr = ndarray_to_mgr(
722 data,
723 index,
724 columns,
725 dtype=dtype,
726 copy=copy,
727 typ=manager,
728 )
730 # For data is list-like, or Iterable (will consume into list)
731 elif is_list_like(data):
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/construction.py:349, in ndarray_to_mgr(values, index, columns, dtype, copy, typ)
344 # _prep_ndarraylike ensures that values.ndim == 2 at this point
345 index, columns = _get_axes(
346 values.shape[0], values.shape[1], index=index, columns=columns
347 )
--> 349 _check_values_indices_shape_match(values, index, columns)
351 if typ == "array":
353 if issubclass(values.dtype.type, str):
File /opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/pandas/core/internals/construction.py:420, in _check_values_indices_shape_match(values, index, columns)
418 passed = values.shape
419 implied = (len(index), len(columns))
--> 420 raise ValueError(f"Shape of passed values is {passed}, indices imply {implied}")
ValueError: Shape of passed values is (4, 3), indices imply (4, 2)
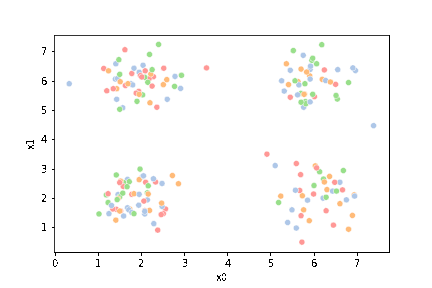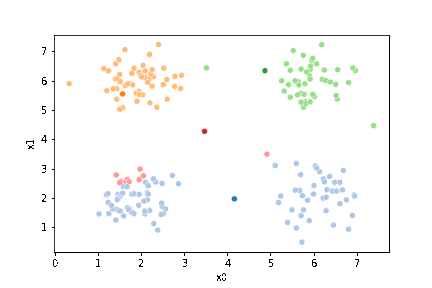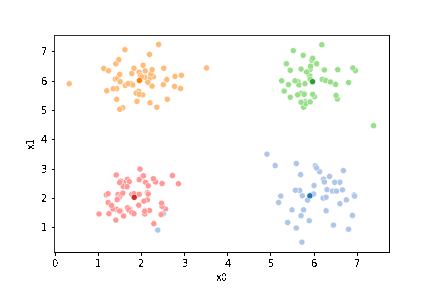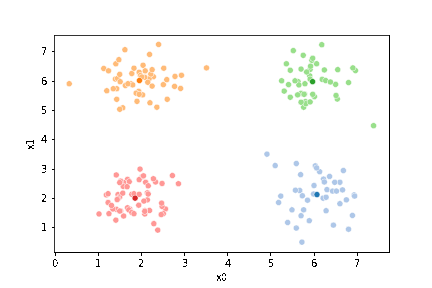
This algorithm is random. So each time we run it, it looks a little different.
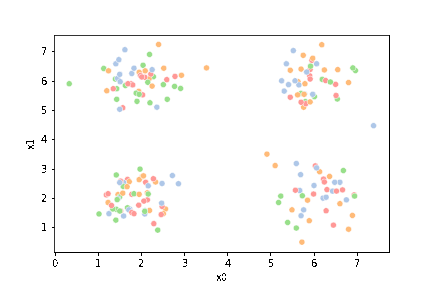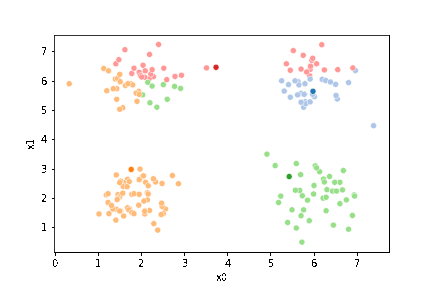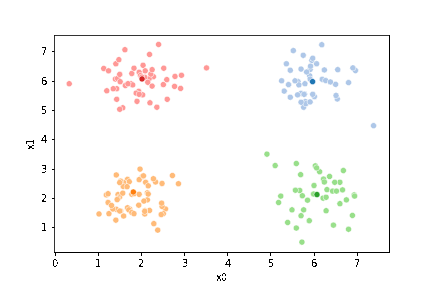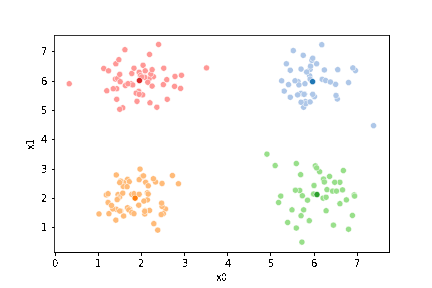
23.3. Questions After class#
23.3.1. Are there any better way to optimize than run multiple times?#
Not totally, the Kmeans ++ algorithm for initialization will help, but sometimes just multiple times is all we have because there is so much more unknown than known.
23.3.2. How do we use sklearn to cluster data?#
We will see that Wednesday, but the short answer is that there is a an estimator object for the clustering model and then we use the fit method