ML Task Review Cross Validation
Contents
31. ML Task Review Cross Validation#
31.1. Relationship between Tasks#
We learned classification first, because it shares similarities with each regression and clustering, while regression and clustering have less in common.
Classification is supervised learning for a categorical target.
Regression is supervised learning for a continuous target.
Clustering is unsupervised learning for a categorical target.
Sklearn provides a nice flow chart for thinking through this.
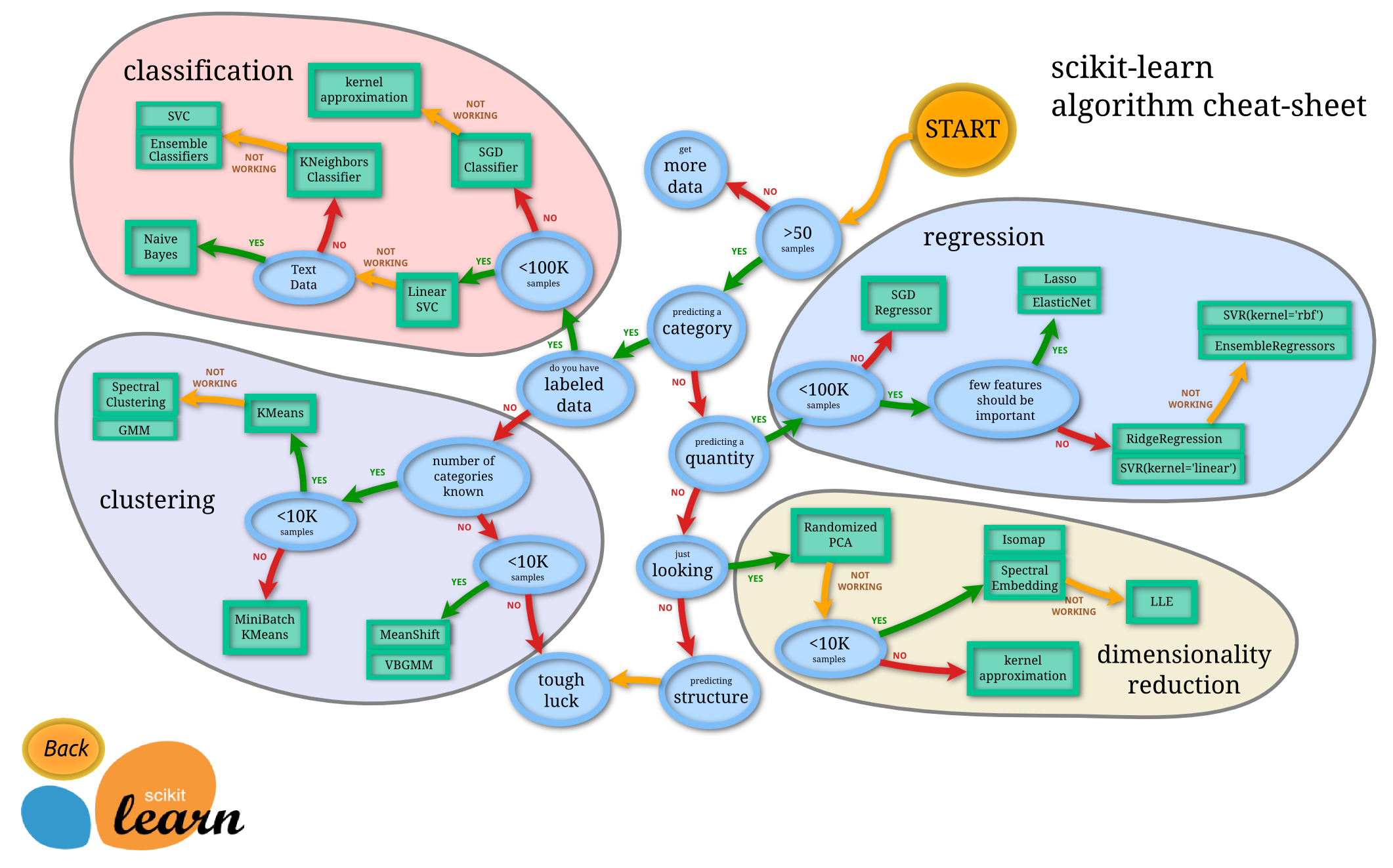
Predicting a category is another way of saying categorical target. Predicting a quantitiy is another way of saying continuous target. Having lables or not is the difference between
The flowchart assumes you know what you want to do with data and that is the ideal scenario. You have a dataset and you have a goal. For the purpose of getting to practice with a variety of things, in this course we ask you to start with a task and then find a dataset. Assignment 9 is the last time that’s true however. Starting with Assignment 10 and the last portflios, you can choose and focus on a specific application domain and then choose the right task from there.
Thinking about this, however, you use this information to move between the tasks within a given type of data. For example, you can use the same data for clustering as you did for classification. Switching the task changes the questions though: classification evaluation tells us how separable the classes are given that classifiers decision rule. Clustering can find other subgroups or the same ones, so the evaluation we choose allows us to explore this in more ways.
Regression requires a continuous target, so we need a dataset to be suitable for
that, we can’t transform from the classification dataset to a regression one.
However, we can go the other way and that’s how some classification datasets are
created.
The UCI adult Dataset is a popular ML dataset that was dervied from census data. The goal is to use a variety of features to predict if a person makes more than \(50k per year or not. While income is a continuous value, they applied a threshold (\)50k) to it to make a binary variable. The dataset does not include income in dollars, only the binary indicator.
Further Reading
Recent work reconsturcted the dataset with the continuous valued income. Their repository contains the data as well as links to their paper and a video of their talk on it.
31.2. Cross Validation#
This week our goal is to learn how to optmize models. The first step in that is to get a good estimate of its performance.
We have seen that the test train splits, which are random, influence the performance.
import pandas as pd
import seaborn as sns
import numpy as np
from sklearn import tree
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import train_test_split
from sklearn.cluster import KMeans
from sklearn import metrics
We’ll use the Iris data with a decision tree.
iris_df = sns.load_dataset('iris')
iris_X = iris_df.drop(columns=['species'])
iris_y = iris_df['species']
dt =tree.DecisionTreeClassifier()
We can split the data, fit the model, then compute a score, but since the splitting is a randomized step, the score is a random variable.
For example, if we have a coin that we want to see if it’s fair or not. We would flip it to test. One flip doesn’t tell us, but if we flip it a few times, we can estimate the probability it is heads by counting how many of the flips are heads and dividing by how many flips.
We can do something similar with our model performance. We can split the data a bunch of times and compute the score each time.
cross_val_score does this all for us.
It takes an estimator object and the data.
By default it uses 5-fold cross validation. It splits the data into 5 sections, then uses 4 of them to train and one to test. It then iterates through so that each section gets used for testing.
cross_val_score(dt,iris_X,iris_y)
array([0.96666667, 0.96666667, 0.9 , 0.93333333, 1. ])
We get back a score for each section or “fold” of the data. We can average those to get a single estimate.
np.mean(cross_val_score(dt,iris_X,iris_y))
0.9533333333333334
We can use more folds.
np.mean(cross_val_score(dt,iris_X,iris_y,cv=10))
0.9533333333333334
We can peak inside what this actually does to see it more clearly.
31.3. What Does Cross validation really do?#
It uses StratifiedKfold for classification, but since we’re using regression it will use KFold. test_train_split uses ShuffleSplit by default, let’s load that too to see what it does.
Warning
The key in the following is to get the concepts not all of the details in how I evaluate and visualize. I could have made figures separately to explain the concept, but I like to show that Python is self contained.
from sklearn.model_selection import KFold, ShuffleSplit
kf = KFold(n_splits = 10)
When we use the split method it gives us a generator.
kf.split(diabetes_X, diabetes_y)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[9], line 1
----> 1 kf.split(diabetes_X, diabetes_y)
NameError: name 'diabetes_X' is not defined
We can use this in a loop to get the list of indices that will be used to get the test and train data for each fold. To visualize what this is doing, see below.
N_samples = len(diabetes_y)
kf_tt_df = pd.DataFrame(index=list(range(N_samples)))
i = 1
for train_idx, test_idx in kf.split(diabetes_X, diabetes_y):
kf_tt_df['split ' + str(i)] = ['unused']*N_samples
kf_tt_df['split ' + str(i)][train_idx] = 'Train'
kf_tt_df['split ' + str(i)][test_idx] = 'Test'
i +=1
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[10], line 1
----> 1 N_samples = len(diabetes_y)
2 kf_tt_df = pd.DataFrame(index=list(range(N_samples)))
3 i = 1
NameError: name 'diabetes_y' is not defined
We can count how many times ‘Test’ and ‘Train’ appear
count_test = lambda part: len([v for v in part if v=='Test'])
count_train = lambda part: len([v for v in part if v=='Train'])
When we apply this along axis=1 we to check that each sample is used in exactly 1 test set how may times each sample is used
sum(kf_tt_df.apply(count_test,axis = 1) ==1)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[12], line 1
----> 1 sum(kf_tt_df.apply(count_test,axis = 1) ==1)
NameError: name 'kf_tt_df' is not defined
and exactly 9 training sets
sum(kf_tt_df.apply(count_test,axis = 1) ==9)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[13], line 1
----> 1 sum(kf_tt_df.apply(count_test,axis = 1) ==9)
NameError: name 'kf_tt_df' is not defined
the describe helps ensure that all fo the values are exa
We can also visualize:
cmap = sns.color_palette("tab10",10)
g = sns.heatmap(kf_tt_df.replace({'Test':1,'Train':0}),cmap=cmap[7:9],cbar_kws={'ticks':[.25,.75]},linewidths=0,
linecolor='gray')
colorbar = g.collections[0].colorbar
colorbar.set_ticklabels(['Train','Test'])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[14], line 2
1 cmap = sns.color_palette("tab10",10)
----> 2 g = sns.heatmap(kf_tt_df.replace({'Test':1,'Train':0}),cmap=cmap[7:9],cbar_kws={'ticks':[.25,.75]},linewidths=0,
3 linecolor='gray')
4 colorbar = g.collections[0].colorbar
5 colorbar.set_ticklabels(['Train','Test'])
NameError: name 'kf_tt_df' is not defined
Note that unlike test_train_split this does not always randomize and shuffle the data before splitting.
If we apply those lambda functions along axis=0, we can see the size of each test set
kf_tt_df.apply(count_test,axis = 0)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[15], line 1
----> 1 kf_tt_df.apply(count_test,axis = 0)
NameError: name 'kf_tt_df' is not defined
and training set:
kf_tt_df.apply(count_train,axis = 0)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[16], line 1
----> 1 kf_tt_df.apply(count_train,axis = 0)
NameError: name 'kf_tt_df' is not defined
We can verify that these splits are the same size as what test_train_split does using the right settings. 10-fold splits the data into 10 parts and tests on 1, so that makes a test size of 1/10=.1, so we can use the train_test_split and check the length.
X_train2,X_test2, y_train2,y_test2 = train_test_split(diabetes_X, diabetes_y ,
test_size=.1,random_state=0)
[len(split) for split in [X_train2,X_test2,]]
Under the hood train_test_split uses ShuffleSplit
We can do a similar experiment as above to see what ShuffleSplit does.
skf = ShuffleSplit(10)
N_samples = len(diabetes_y)
ss_tt_df = pd.DataFrame(index=list(range(N_samples)))
i = 1
for train_idx, test_idx in skf.split(diabetes_X, diabetes_y):
ss_tt_df['split ' + str(i)] = ['unused']*N_samples
ss_tt_df['split ' + str(i)][train_idx] = 'Train'
ss_tt_df['split ' + str(i)][test_idx] = 'Test'
i +=1
ss_tt_df
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[17], line 2
1 skf = ShuffleSplit(10)
----> 2 N_samples = len(diabetes_y)
3 ss_tt_df = pd.DataFrame(index=list(range(N_samples)))
4 i = 1
NameError: name 'diabetes_y' is not defined
And plot
cmap = sns.color_palette("tab10",10)
g = sns.heatmap(ss_tt_df.replace({'Test':1,'Train':0}),cmap=cmap[7:9],cbar_kws={'ticks':[.25,.75]},linewidths=0,
linecolor='gray')
colorbar = g.collections[0].colorbar
colorbar.set_ticklabels(['Train','Test'])
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[18], line 2
1 cmap = sns.color_palette("tab10",10)
----> 2 g = sns.heatmap(ss_tt_df.replace({'Test':1,'Train':0}),cmap=cmap[7:9],cbar_kws={'ticks':[.25,.75]},linewidths=0,
3 linecolor='gray')
4 colorbar = g.collections[0].colorbar
5 colorbar.set_ticklabels(['Train','Test'])
NameError: name 'ss_tt_df' is not defined
31.4. Cross validation with clustering#
We can use any estimator object here.
km = KMeans(n_clusters=3)
cross_val_score(km,iris_X)
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
array([ -9.062 , -14.93195873, -18.93234207, -23.70894258,
-19.55457726])
km.score()
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[21], line 1
----> 1 km.score()
TypeError: score() missing 1 required positional argument: 'X'
31.5. Grid Search Optimization#
We can optimize, however to determing the different parameter settings.
A simple way to do this is to fit the model for different parameters and score for each and compare.
The
from sklearn.model_selection import GridSearchCV
The GridSearchCV object is constructed first and requires an estimator object and a dictionary that describes the parameter grid to search over.
The dictionary has the parameter names as the keys and the values are the values for that parameter to test.
The fit method on the Grid Search object fits all of the separate models.
In this case, we will optimize the depth of this Decision Tree.
param_grid = {'max_depth':[2,3,4,5]}
dt_opt = GridSearchCV(dt,param_grid)
dt_opt.fit(iris_X,iris_y)
GridSearchCV(estimator=DecisionTreeClassifier(),
param_grid={'max_depth': [2, 3, 4, 5]})In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(estimator=DecisionTreeClassifier(),
param_grid={'max_depth': [2, 3, 4, 5]})DecisionTreeClassifier()
DecisionTreeClassifier()
Then we can look at the output.
dt_opt.cv_results_
{'mean_fit_time': array([0.00185766, 0.001791 , 0.00178885, 0.00179324]),
'std_fit_time': array([1.59000176e-04, 2.61296321e-05, 1.73072840e-05, 2.34260540e-05]),
'mean_score_time': array([0.00122561, 0.0011929 , 0.00116849, 0.00119224]),
'std_score_time': array([6.78506939e-05, 2.81352511e-05, 1.85231443e-05, 3.18513659e-05]),
'param_max_depth': masked_array(data=[2, 3, 4, 5],
mask=[False, False, False, False],
fill_value='?',
dtype=object),
'params': [{'max_depth': 2},
{'max_depth': 3},
{'max_depth': 4},
{'max_depth': 5}],
'split0_test_score': array([0.93333333, 0.96666667, 0.96666667, 0.96666667]),
'split1_test_score': array([0.96666667, 0.96666667, 0.96666667, 0.96666667]),
'split2_test_score': array([0.9 , 0.93333333, 0.9 , 0.9 ]),
'split3_test_score': array([0.86666667, 0.93333333, 0.93333333, 0.96666667]),
'split4_test_score': array([1., 1., 1., 1.]),
'mean_test_score': array([0.93333333, 0.96 , 0.95333333, 0.96 ]),
'std_test_score': array([0.04714045, 0.02494438, 0.03399346, 0.03265986]),
'rank_test_score': array([4, 2, 3, 1], dtype=int32)}
We note that this is a dictionary, so to make it more readable, we can make it a DataFrame.
pd.DataFrame(dt_opt.cv_results_)
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_max_depth | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.001858 | 0.000159 | 0.001226 | 0.000068 | 2 | {'max_depth': 2} | 0.933333 | 0.966667 | 0.900000 | 0.866667 | 1.0 | 0.933333 | 0.047140 | 4 |
| 1 | 0.001791 | 0.000026 | 0.001193 | 0.000028 | 3 | {'max_depth': 3} | 0.966667 | 0.966667 | 0.933333 | 0.933333 | 1.0 | 0.960000 | 0.024944 | 2 |
| 2 | 0.001789 | 0.000017 | 0.001168 | 0.000019 | 4 | {'max_depth': 4} | 0.966667 | 0.966667 | 0.900000 | 0.933333 | 1.0 | 0.953333 | 0.033993 | 3 |
| 3 | 0.001793 | 0.000023 | 0.001192 | 0.000032 | 5 | {'max_depth': 5} | 0.966667 | 0.966667 | 0.900000 | 0.966667 | 1.0 | 0.960000 | 0.032660 | 1 |
dt
DecisionTreeClassifier()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier()
31.6. Questions After Class#
31.6.1. Do we have to do anything to pick the highest ranking model from the GridSearchCV function?#
No, we can use it directly. For example:
plt.figure(figsize=(15,20))
tree.plot_tree(dt_opt.best_estimator_, rounded =True, class_names = ['A','B'],
proportion=True, filled =True, impurity=False,fontsize=10);
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[28], line 1
----> 1 plt.figure(figsize=(15,20))
2 tree.plot_tree(dt_opt.best_estimator_, rounded =True, class_names = ['A','B'],
3 proportion=True, filled =True, impurity=False,fontsize=10);
NameError: name 'plt' is not defined
31.6.2. Is there anything similar to a gridsearch object for different estimators, where it can try different methods of estimation and rank them?#
No, you would use multiple gridsearch (or similar model optimizer with a different search strategy) one for each model. Each model class/ estimator object
31.6.3. I would like to learn how to apply cross validation and especially program optimization to unsupervised clustering models.#
It would look a lot like what we did with the decision tree, but we use the right parameter name, for example:
km = KMeans()
param_grid = {'n_clusters': list(range(2,8))}
km_opt = GridSearchCV(km,param_grid)
km_opt.fit(iris_X)
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/opt/hostedtoolcache/Python/3.9.16/x64/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
GridSearchCV(estimator=KMeans(), param_grid={'n_clusters': [2, 3, 4, 5, 6, 7]})In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(estimator=KMeans(), param_grid={'n_clusters': [2, 3, 4, 5, 6, 7]})KMeans()
KMeans()
pd.DataFrame(km_opt.cv_results_)
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_n_clusters | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.007247 | 0.002357 | 0.001162 | 0.000065 | 2 | {'n_clusters': 2} | -13.417292 | -19.334754 | -58.409800 | -56.347656 | -54.802470 | -40.462394 | 19.788398 | 6 |
| 1 | 0.008210 | 0.000141 | 0.001154 | 0.000030 | 3 | {'n_clusters': 3} | -9.062000 | -14.931959 | -18.932342 | -23.708943 | -19.554577 | -17.237964 | 4.945204 | 5 |
| 2 | 0.009823 | 0.000376 | 0.001202 | 0.000036 | 4 | {'n_clusters': 4} | -9.062000 | -11.602296 | -13.714817 | -17.596427 | -15.673617 | -13.529831 | 2.994800 | 4 |
| 3 | 0.011454 | 0.000734 | 0.001220 | 0.000014 | 5 | {'n_clusters': 5} | -9.062000 | -10.723786 | -10.996277 | -17.596427 | -15.673617 | -12.810421 | 3.249602 | 3 |
| 4 | 0.012644 | 0.000161 | 0.001230 | 0.000007 | 6 | {'n_clusters': 6} | -9.062000 | -8.024337 | -10.996277 | -12.698270 | -11.476818 | -10.451540 | 1.686290 | 2 |
| 5 | 0.013671 | 0.000281 | 0.001206 | 0.000021 | 7 | {'n_clusters': 7} | -9.062000 | -7.158558 | -9.066973 | -12.183027 | -11.476818 | -9.789475 | 1.819295 | 1 |
31.6.4. Is it better to split the data in more folds when using the cross-validation?#
this is a tricky question, we’ll revisit it in class on Wednesday.
31.6.5. ‟What is this “model” we are training? What are the scores, scoring?#
In this example, the model was a decision tree at the beginning and later K-means. The score describes how well the fit model works on the held out data; accuracy or a general fit statistic.
The model vs algorithm section in the introduction of the Model Based ML book (free) is a good thing to read to clarify these relationshipts.
sklearn provides a flowchart for choosing their different estimator objects. In sklearn, they implement each model as an estimator object; more specifically, they have a Base Estimator class that the other estimators inherit. For example the decision tree source shows that it inherits the ClassifierMixin and BaseDecisionTree which inherits BaseEstimator
Term |
Definition |
Example |
task |
the type of algorithm that we will use machine learning to write |
classification, regression, clustering |
model |
the specific form and set of assumptions that will be used in the algorithm |
decision tree (classification) Gaussian Naive Bayes (classification), linear regression, sparse regression/LASSO, K-means, spectral clustering, etc. |
score |
a measure of how well the model completes the task |
accuracy (for classification), mean squared error (for regression), silhouette score (for clustering) |
also review the intro to models in machine learning class notes